Specificity quantitive detection for cell apoptosis
A cell and detection technology, applied in the field of cell biology, can solve time-consuming and labor-intensive problems, achieve reliable guiding principles, high specificity, and improve the effect of accurately detecting the occurrence of apoptosis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0075] Example 1 Selective labeling test for fragmented DNA fragments in apoptosis
[0076] In early apoptotic cells, the cell membrane remains intact, but the cells are rapidly phagocytized by macrophages or surrounding cells before being lysed under lysis. However, necrosis, another cell death process, is typically caspase-independent and has typical features of increased membrane permeability and enlarged cells and organelles. Therefore, this embodiment makes full use of the membrane defect characteristics of necrotic cells to achieve the purpose of specifically identifying apoptotic cells.
[0077] The specific method is: the 5X10 3 / well of HT-29 cells at 37 °C with H 2 o 2 Treat for one hour to induce cell necrosis; then wash with PBS and incubate in medium containing 10% FCS (fetal calf serum). Live 5X10 3 / well of HT-29 cells was then added to the H 2 o 2 In the induced necrotic cell group, cells were subsequently treated with 5Gy of γ-irradiation or 100nM paclita...
Embodiment 2
[0079] Example 2 Detection of cell apoptosis in HT-29 cells
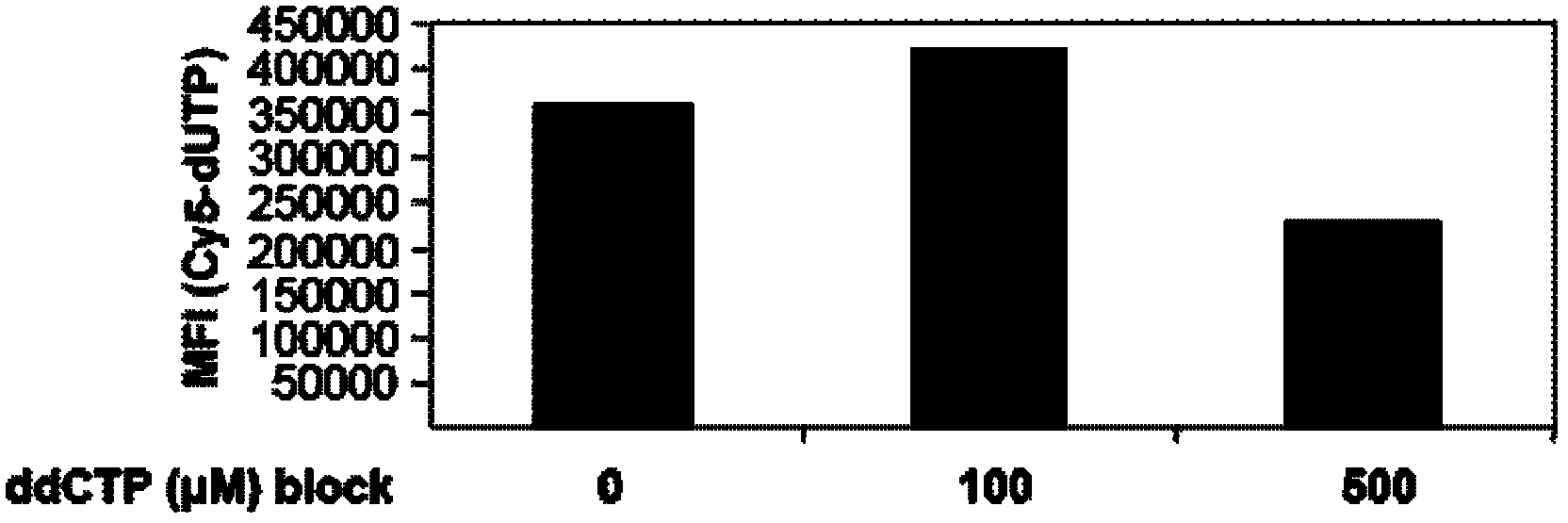
[0080] Pre-incubation of cells with unlabeled ddCTP is necessary to distinguish non-specific DNA fragmentation from apoptosis-induced DNA fragmentation.
[0081] In order to test the hypothesis of the present invention that pre-incubation of unlabeled ddCTP can block the detection of pre-formed DNA breaks, terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling assay (TUNEL) can be performed first. That is to say, after the cells are treated with DNase I (HT-29 cells are used in the present invention), DNA fragmentation can be generated in the cells, and the DNA caused by this non-apoptosis will be detected by the conventional TUNEL apoptosis detection technology. fracture. However, this false positive rate was significantly reduced after pre-incubation with ddCTP.
[0082] The specific operation is as follows: HT-29 cells are placed in 4-well glass slides (10000 cells / well), first incubated with 10 ...
Embodiment 3
[0083] Example 3 Detection of Apoptosis in Tumor Cells
[0084] To demonstrate that this technique can also be used to detect specific DNA breaks in tumor biopsies, tumor tissue sections were embedded on glass slides (including necrotic cells), and the tissue sections were first processed as follows:
[0085] 1. Dewaxing: xylene dewaxing twice, 5-10 minutes each time; ethanol hydration (put the dewaxed slices into 100% ethanol, 95% ethanol, 90% ethanol, 80% ethanol, 70% ethanol ethanol for 2 minutes each)
[0086] 2. Immerse the slices in PBS and rinse 3 times, 5 minutes each time.
[0087] 3. Lay the slices horizontally, add proteinase K working solution on the tissue block, and act for 15-30 minutes at 21-37°C. (Preparation of Proteinase K working solution: 2μL 50×Protease K+98μL PBS)
[0088] 4. Immerse the proteinase K-treated sample in PBS and rinse 3 times, 5 minutes each time.
[0089] 5. Immerse the slices in the blocking solution and block at room temperature (15-...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com