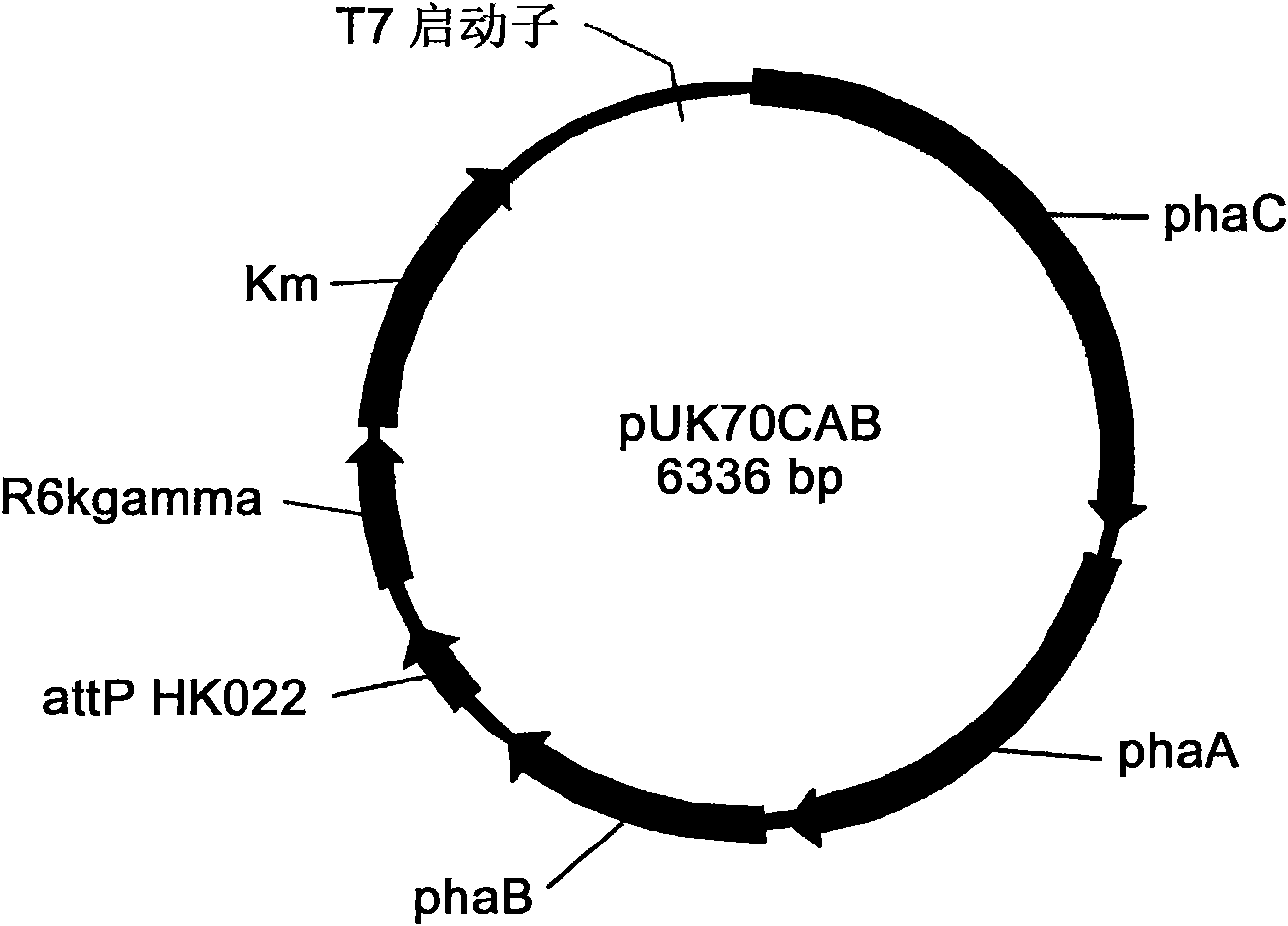
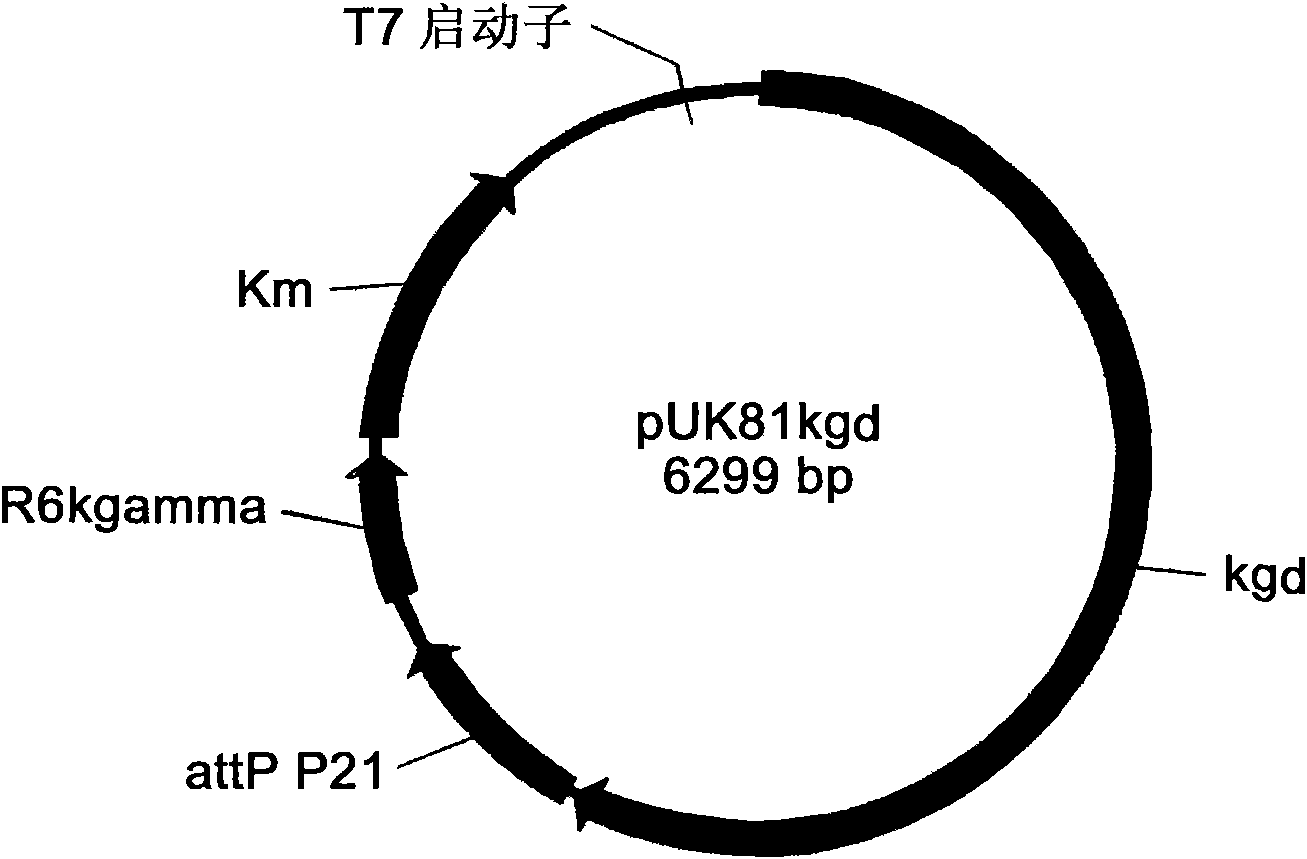
Engineering bacterium containing 2-oxoglutarate decarboxylase gene kgd and applications thereof
A technology of engineering bacteria and genes, applied in the field of genetic engineering technology and fermentation, can solve the problems of high price, high cytotoxicity, cumbersome and complicated mixed carbon source fermentation strategy, etc., and achieve the effect of reducing production costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Embodiment 1, construction Escherichia coli genome integration plasmid pKD13km
[0040] 1. Digest the plasmid pKD13 with AceIII, recover the 2.5kb fragment by gel, and obtain the fragment containing the Km resistance gene and the R6kgamma replicon.
[0041] 2. Use the solutionI ligation kit of Takara (the solutionI kit is T4DNA ligase, and the product number in Takara company is D6022) to carry out self-ligation reaction to the fragment recovered in step 1 (the fragment and solutionI ratio after digestion are 1: 1), the ligation product was introduced into E.coli S17-1(λpir) by electroporation (Herrero, M, V.deLorenzo, and K.N.Timmis.1990.Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable Chromosomal insertion of foreign genes in gram-negative bacteria.J.Bacteriol.172:6557-6567), spread on LB-Km solid medium, and culture at 37°C for 16h.
[0042] 3. Plasmid verification: Pick a single clone from the LB-Km solid medium and p...
Embodiment 2
[0043] Example 2. Construction of genome integration expression vectors pUK series pUK162, pUK70, pUK63, pUK81 and pUK154.
[0044] 1. Under standard polymerase chain reaction conditions, use pEASY-Blunt as a template, primer P1: CCATCGCCCTGATAGACGGT (SalI) and primer P2: ACCTTTCCTTTTTCAATTCAGAAG (EcoRI) as upstream and downstream primers to amplify a 1365bp DNA fragment. Sequencing showed that this fragment contained the Km resistance gene. The Km resistance gene fragment was recovered after treatment with endonucleases EcoRI and SalI.
[0045] 2. Under standard polymerase chain reaction conditions, using pEASY-Blunt as a template, using primer P3: ATAAGAATTCGGCAACTATGGATGAACGAA (EcoR) and primer P4: ATATGTCGACTAATACGACTCACTATAGGGCGA (SalI) as upstream and downstream primers to amplify a 1409bp DNA fragment. Sequencing showed that the fragment contained the replicon sequence of pEASY-Blunt, and the replicon gene fragment was recovered after treatment with endonucleases EcoRI...
Embodiment 3
[0068] Embodiment 3, construction plasmid pBHR68orfZ
[0069] 1. Digest plasmid pCK3 with ClaI and EcoRI ( B, Gottschalk G. Molecular analysis of the anaerobic succinate degradation pathway in Clostridium kluyveri. J Bacteriol, 1996, 178: 871-880), a 1.8 kb fragment was recovered from the gel, which contained the orfZ gene and its promoter sequence.
[0070]2. Digest plasmid pBHR68 with ClaI and EcoRI (Spiekermann P, Rehm BHA, Kalscheuer R, et al. A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch Microbiol, 1999, 171: 73-80), the gel recovered a fragment of 8.1 kb, containing phaCAB gene and its promoter sequence, Amp resistance gene and replicon derived from pBluescript II SK (-).
[0071] 3. Use T4 DNA ligase to connect the fragments recovered in step 1 and step 2; after the ligation reaction is completed, the plasmid pBHR68orfZ is obtained, and the ob...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com