PCR (Polymerase Chain Reaction) method and kit for quickly detecting Enterobacter sakazakii in baby formula
A technology of Enterobacter sakazakii and infant formula, which is applied in the fields of biochemical equipment and methods, microbe determination/inspection, and resistance to vector-borne diseases, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0083] Kit composition and preparation
[0084] 1. DNA extraction solution: including the following components: 10×TE (0.1M Tris-HCL, 0.1M EDTA pH8.8); 50ug / ul lysozyme; 1% SDS; 0.2ug / ul proteinase K.
[0085] 2. 10×PCR reaction solution: including 100mM KCL; 1200mM Tris-HCL; 1% Triton x-100; 15mM MgSO 4 ; 100mM (NH 4 ) 2 SO 4 ; 1 mg / mL BSA;
[0086] 3. Standard positive template: The standard positive template is composed of pGEMT-easy vector containing a 265-base nucleotide fragment of the conserved gene ompA of Enterobacter sakazakii.
[0087] 4. Specific primers and molecular beacons:
[0088] 5-ACCGGCGTTTCTCCTGTA-3
[0089] 5-TCAGCATGCCGTTGTCC-3
[0090] Fluorescence signal FAM-ctgcat-CGGGGTTTCTCCTGTATTCG-atgcag-DABCYL quenches the signal.
[0091] 5. Internal standard recombinant plasmid
[0092] The standard internal standard control is a pUC18T recombinant plasmid constructed from the artificially synthesized sequence containing the human MICA gene and the Mol...
Embodiment 2
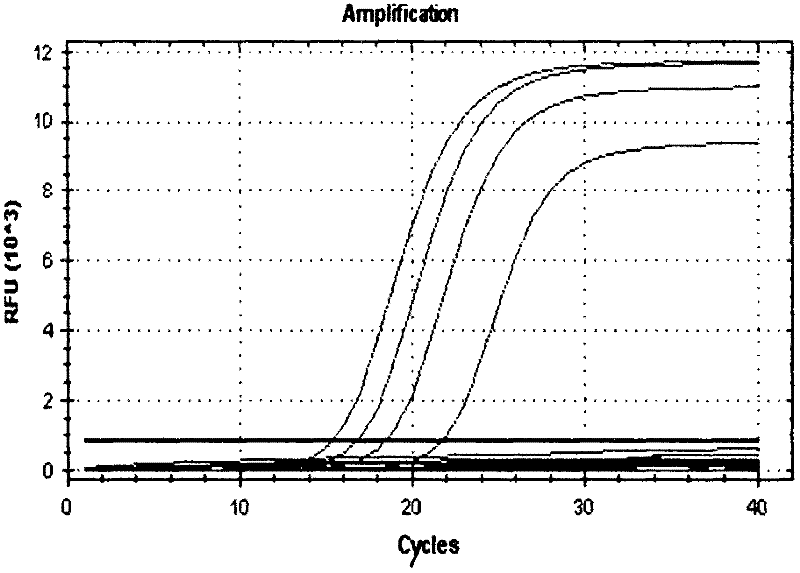
[0096] Embodiment 2: the sensitivity test of kit
[0097] 1. Culture of Enterobacter sakazakii
[0098] Standard strains of Enterobacter sakazakii were purchased and cultured overnight in bacterial broth for later use.
[0099] 2. Preparation of Enterobacter sakazakii genomic DNA.
[0100] After the strain was cultured overnight in the bacterial broth, the bacteria were suspended in 500ul TE buffer, added with a final concentration of 50ug / ul lysozyme, and reacted at 37°C for 1 hour, then added a final concentration of 1% SDS and 0.2ug / ul proteinase K, After 1 h at 55°C, the DNA was extracted with phenol-chloroform.
[0101]3. PCR amplification conditions: PCR reaction system (25 μL): 2 μL of 10×PCR buffer, 1 μL of each primer pair and molecular beacon (10 μmol / L), 2 μL of dNTPs (10 mmol / L), TaqDNA polymerase (5 U / μL ) 0.2 μL, template DNA 2 μL, water 16.8 μL. PCR reaction conditions: 94°C for 3min; 94°C for 60s, 60°C for 60s, 40 cycles; observe the amplification curve in ...
Embodiment 3
[0102] Embodiment 3: the specificity test of kit
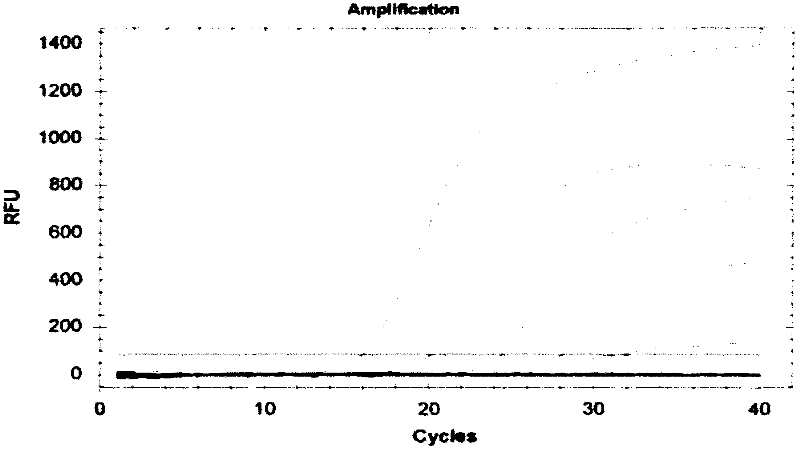
[0103] Salmonella schottmuelleri; Salmonella pafatyphi B; Salmonella typhimurium; Salmonella enreritidis; Shigella sonnei; Shlgella dysenteriae; Shigella boydii; Shigella flexnei; Enterobacteo yersinia); Proteus vuLgaris; Listeria monocytogenes; Escherichia coli (Eseheriehiaeoli) as a control, and Enterobacter sakazakii ATCC29544 strain; ATCC 51007 strain; ATCC 51024 strain DNA, positive control plasmid, internal standard recombinant plasmid, each take 5uL as a template for PCR reaction. At the same time, a negative control was set up.
[0104] The PCR reaction conditions were: PCR reaction system (25 μL): 2 μL of 10×PCR buffer, 1 μL of each primer pair and molecular beacon (10 μmol / L), 2 μL of dNTPs (10 mmol / L), 0.2 μL of Taq DNA polymerase (5 U / μL) μL, template DNA 2 μL, water 16.8 μL.
[0105] PCR reaction conditions: 94°C for 3min; 94°C for 60s, 60°C for 60s, 40 cycles.
[0106] Results Only Enterobacter sakazakii (En...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com