Application of LeEXP2 to improving efficiency of cellulase to degrade cellulosic materials
A technology for degrading cellulose and cellulase, applied in the field of bioengineering, can solve problems such as cumbersome steps of Expansin protein, difficult expression and purification of plant expansin, and cooperation with cellulase to improve cellulose degradation efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Cloning of embodiment 1 tomato LeEXP2 gene
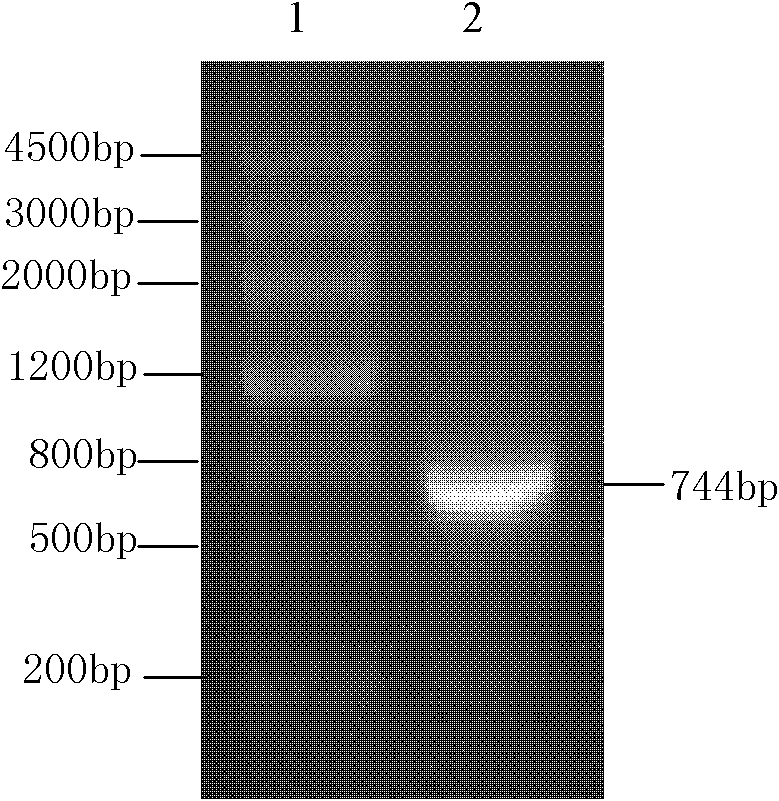
[0034] Use the Trizol reagent to extract the total RNA of tomato according to the kit instructions: get 2ug of total RNA, use the reverse transcription kit to reverse-transcribe it into cDNA (Promega Company), use the cDNA as a template, and use the SEQ ID NO. 1 and the nucleotide sequence shown in SEQID NO.2 carry out PCR for upstream primer and downstream primer, amplify the LeEXP2 gene of 744bp shown in SEQ ID NO.3 in the sequence listing (see figure 1 ). The PCR product was recovered and connected to the TA carrier pGEM-T (Promega Company) to obtain a new plasmid, which was confirmed by sequencing to contain the tomato LeEXP2 gene, and the plasmid was named pGEM-LeEXP2, that is, the plasmid pGEM-LeEXP2 containing the tomato LeEXP2 gene was obtained. build strategy see figure 2 .
Embodiment 2
[0035] Example 2 Construction of LeEXP2 gene to Pichia pastoris expression vector
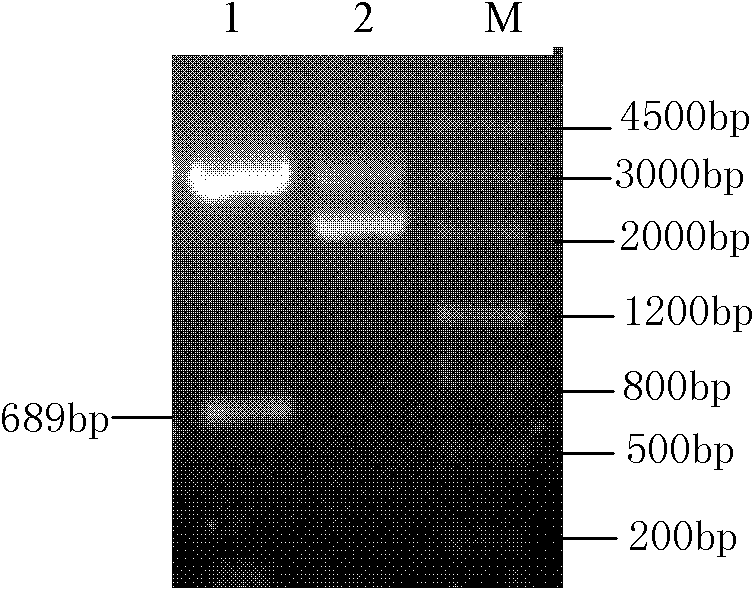
[0036] Using the plasmid pGEM-LeEXP2 as a template, using the nucleotide sequence shown in SEQ ID NO.4 in the sequence listing as an upstream primer, and using the nucleotide sequence shown in SEQ ID NO.5 in the sequence listing as a downstream primer for PCR amplification, Obtain the 703bp fragment shown in SEQID NO.6 in the sequence listing, and connect the 703bp fragment with the TA cloning vector pGEM-T to transform the transformant obtained from Escherichia coli TOP10F'. It has been identified that the plasmid in the transformant is composed of LeEXP2 gene and pGEM -T connection, the new plasmid obtained from the transformant was named pTLeEXP2, and the plasmid pTLeEXP2 was extracted from the above-mentioned transformant, and the pPICZalpha A plasmid was extracted from Escherichia coli containing the yeast expression vector pPICZalpha A plasmid, and simultaneously enzyme Plasmids pTLeEXP2 ...
Embodiment 3
[0037] Example 3 Integration of LeEXP2 gene into Pichia pastoris chromosome:
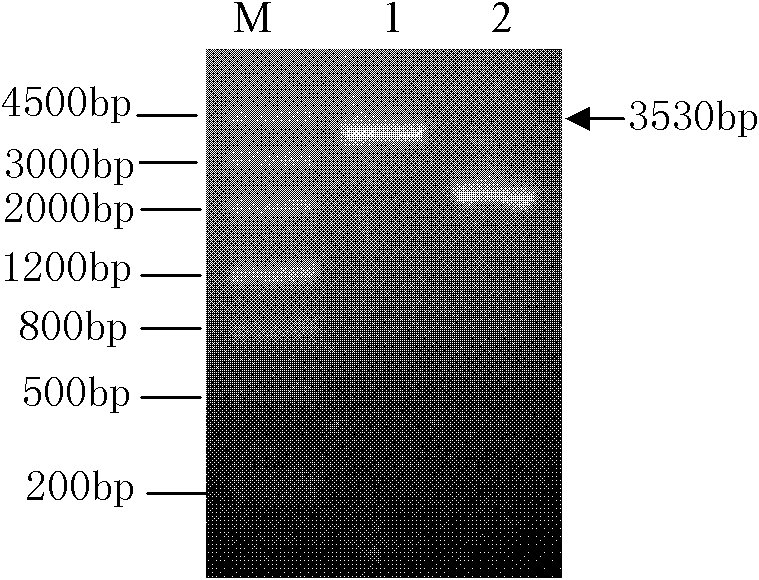
[0038] Take 10-15 μg of pPICZalpha A-LeEXP2 plasmid, use PmeI enzyme to cut into the 4219bp fragment shown in the linear SEQ ID NO.9, electrophoresis detection is as follows Figure 7 A 4219bp fragment is generated as shown. After extraction with phenol / chloroform, ethanol precipitates the linear DNA fragment shown in the linear SEQ ID NO.9. After drying, it is dissolved in sterile water to make a linear DNA solution with a DNA concentration of 0.5-1μg / μL ;
[0039] Prepare the competent cells of Pichia pastoris host strain X-33 by referring to the electroporation transformation preparation yeast competent cell method in the refined Molecular Biology Experiment Guide (Fourth Edition) (P512-513), and 80 μL Bath The competent cells of Pichia host strain X33 were mixed with 10 μL of 0.5-1 μg / μL linear DNA solution, then transferred to a pre-cooled electrode cup at 4 °C, ice-bathed for 5 min, and the v...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com