Molecular marker closely linked with salt-tolerant main effect QTL (Quantitative Trait Loci) at tomato seedling stage and application thereof
A molecular marker, tomato seedling technology, applied in the field of agricultural biology, can solve the problems of slow progress of tomato salt tolerance breeding, complex tomato salt tolerance genetic mechanism, long breeding cycle, etc., so as to improve the accuracy of selection and breeding efficiency, improve the Breeding and selection efficiency, production cost savings
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Example 1: Acquisition of Molecular Markers Tightly Linked to Salt Tolerance Major QTL in Tomato Seedling Stage
[0052] (1) F 1 mongrel.
[0053] (2)F 1 Selfing produces 1338 F 2 Individual plants, the genomic DNA of each F2 individual plant was isolated, and 1338 F2 strains were screened using the flanking marker SSR285 on the salt-tolerant subintrogression line IL7-5-5. 2 single plant, a 255-strain F 2 Regroup.
[0054] (3) According to existing methods (Song Jian, Journal of Shanghai Jiaotong University (Agricultural Science Edition), 2006, 24(6): 524-528; Wu Yuhui, China Agricultural Science Bulletin, 2008, 24(4): 80-84; Lei Na, Journal of Northeast Agricultural University, 2008, 39(3): 29-33), isolated the DNA of parental IL7-5-5 and M82, and used microsatellite (SSR), enzyme-cut amplified polymorphic sequence (CAPS) and Amplified fragment length polymorphism (AFLP) primers were used to perform PCR amplification on the two parents, and the amplified products...
Embodiment 2
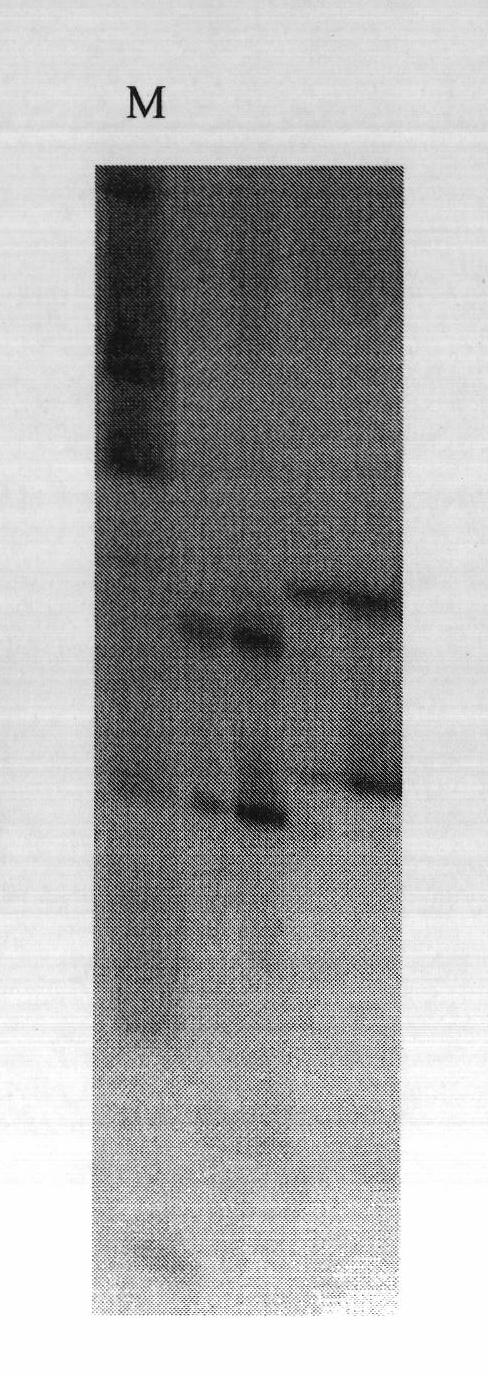
[0065] According to the existing technology, adopt: Song Jian, Journal of Shanghai Jiaotong University (Agricultural Science Edition), 2006, 24(6): 524-528; Wu Yuhui, China Agricultural Science Bulletin, 2008, 24(4): 80-84; Lei Na , Journal of Northeast Agricultural University, 2008, 39(3): 29-33, selecting primer combinations of SSR, CAPS and AFLP with amplified polymorphism between the two parents IL7-5-5 and M82. After PCR amplification, the amplified products were separated by electrophoresis on 1.5% agarose gel and 6% denaturing polyacrylamide gel, and the polymorphic markers obtained were respectively SSR primers SSR285, CAPS Mark U231219 and C 2 - At4g30580, AFLP markers E35 / M49 and E39 / M50.
[0066] The primer sequences used were:
[0067] The upstream primer of SSR285 is 5'AGTGGCTCTCACCTACTGCG'3, and the downstream primer is 5'CAATTCTCAGGCATGAAACG'3;
[0068] The upstream primer of U231219 is 5'AGTCTCTTGCTTGAGTCTGGAGTTG'3, and the downstream primer is 5'TGCTGGGAATA...
Embodiment 3
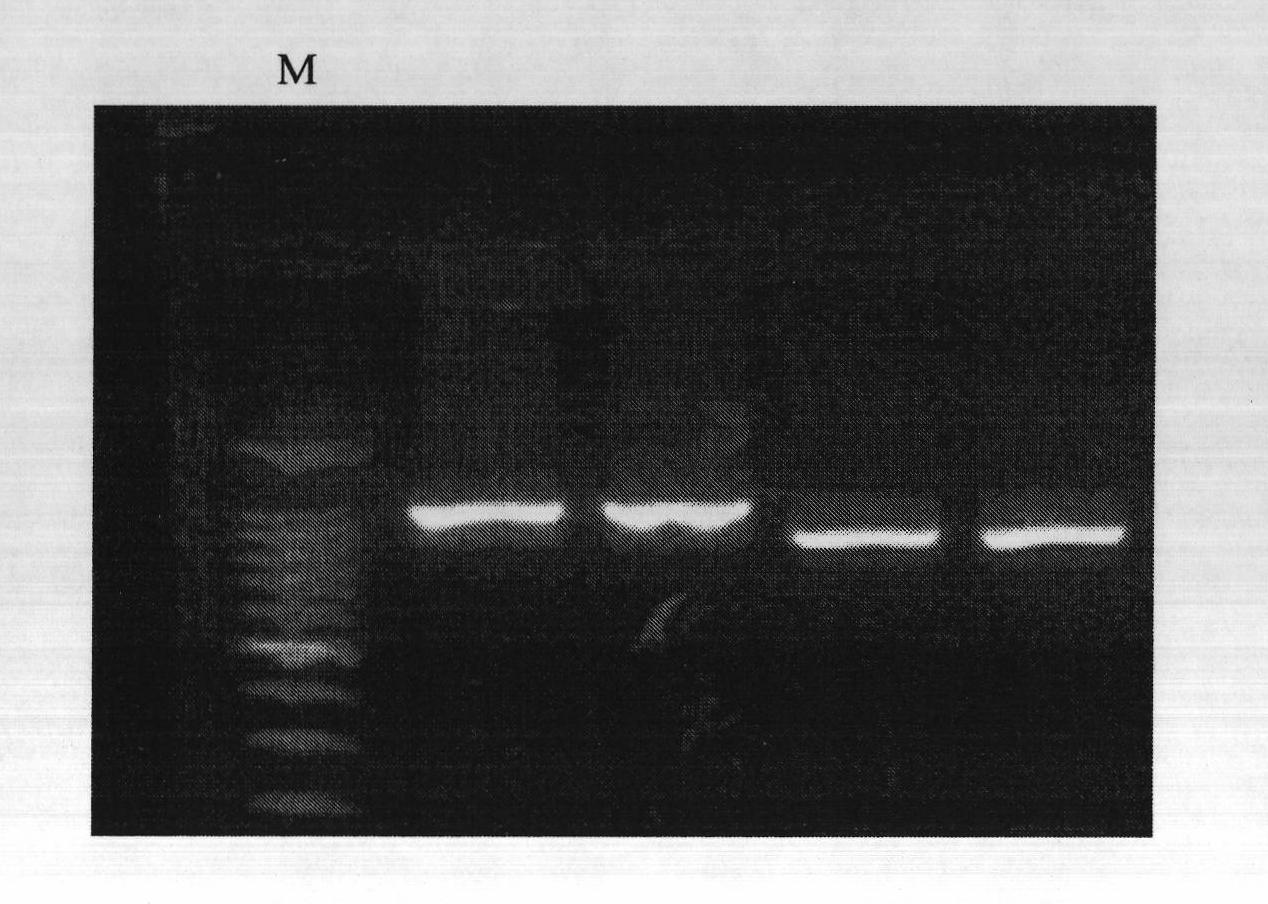
[0073] Example 3: Molecular markers closely linked to main effect QTL for salt tolerance in tomato seedling stage are in F 2 Validation in segregated populations
[0074] The obtained molecular markers closely linked to the major QTL for salt tolerance in tomato seedling stage were found in the F 2 Part of the plants of the first generation were predicted for seedling salt tolerance. DNA was isolated from their leaves, and then the DNA was amplified by PCR using primers labeled SSR285, C2_At4g30580, U231219, E35 / M49 and E39 / M50, and passed Judging the map to determine whether there are corresponding markers, if there are more than 3 molecular markers on the opposite side of the main QTL for salt tolerance, it means that the line belongs to the salt-tolerant line at the seedling stage, and if it does not exist, it is a salt-sensitive material. Then based on these judgment criteria, the seedling salt tolerance of each tomato plant was predicted. Subsequently, the actual salt t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com