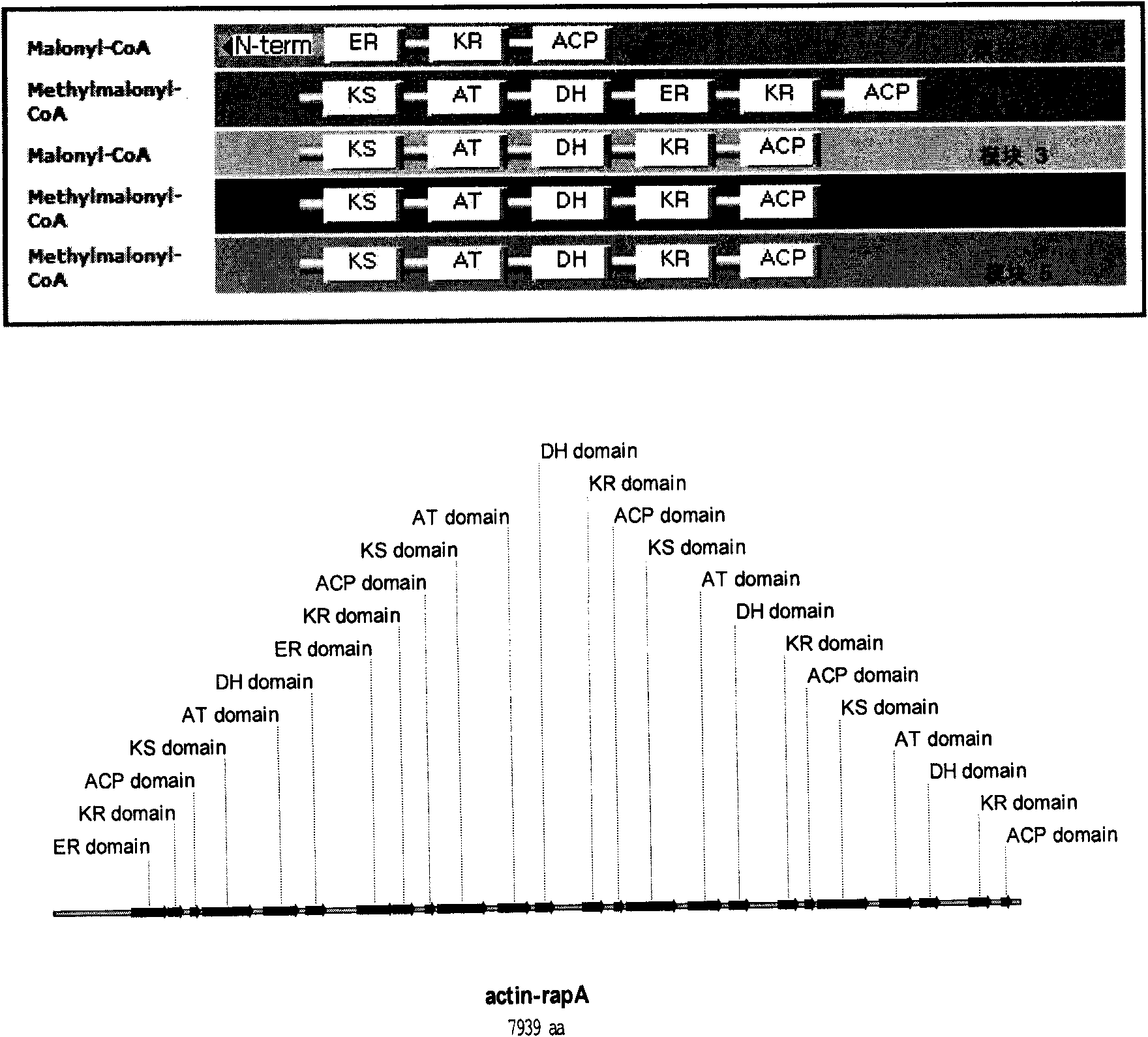
Rapamycin biosynthesis gene from actinoplanes, and separation method and application thereof
A technology of actinomycetes mobilis and rapamycin, which is applied in the field of genetic engineering and can solve problems such as unreported research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Extraction of total DNA from rapamycin producing bacteria Actinomycetes mobilis
[0043] Source of bacteria
[0044]Actinomycetes mobilis is consistent with the literature source: Nishida, H., T. Sakakibara, et al. (1995). "Generation of novel rapamycin structures by microbial manipulations." J Antibiot (Tokyo) 48 (7): 657 -66.
[0045] Inoculate a small amount of actinomycete mycelium into a test tube containing 5ml YEME medium, 30°C, shake culture for about 72 hours, inoculate 5% to 50ml YEME medium, 30°C, shake culture for 24 hours, centrifuge at 3500rpm The cell phone mycelium was washed twice with lysate and stored at -20°C for the extraction of genomic DNA. Add 10ml of lysate (containing lysozyme 5mg / ml) to the mycelium of the mobile phone, vortex until uniform, bathe in 37°C water bath for 60 minutes, add 0.1ml proteinase K (4mg / ml), 1ml 10% SDS, mix well Quickly put it into a 55°C water bath for 60 minutes until the solution is clarified. Cool on ice, add 2....
Embodiment 2
[0047] Construction of Genomic Library of Rapamycin-producing Bacteria
[0048] Firstly, the total DNA of Actinomycetes mobilis obtained in Example 1 was mechanically disrupted to obtain a DNA fragment with a size of 20-40 kb, which was recovered. Escherichia coli EPI100 was streaked on a blank LB plate and cultivated overnight at 37°C. Pick EPI100 monoclonal and inoculate in 3mlLB (containing 10mM MgSO 4 ) in liquid medium, shake the bacteria at 37°C until OD 600 =0.8-1.0 and set aside at 4°C. The total DNA size of 20-30kb fragments were ligated to the fosmid vector overnight. Take out the packaging mixture (containing 50ul of packaged protein) from the -80°C refrigerator and place it in a dry ice bucket. Melt the packaged material immediately. Take out 25ul of the packaged protein and put it in a 1.5ml centrifuge tube and add 6ul of the ligated product just after it starts to melt. , and mix well in a 30°C water bath for 90 minutes. The remaining 25ul of packaged protei...
Embodiment 3
[0050] Nucleic acid molecular hybridization
[0051] 1. DIG DNA labeling: Dilute the DNA to be labeled with sterile water to a total volume of 15ul, heat and denature in boiling water for 10 minutes, and immediately cool in ice. Then add 2ul of primer mixture, 2ul of dNTP mixture, and 1ul of enzyme, mix well, and put in a 37°C water bath for about 16 hours. Add 0.8ul 0.8M EDTA (pH8.0) to terminate the reaction, add 2.5ul 4M LiCl and mix well, then add 75ul pre-cooled absolute ethanol to precipitate the labeled DNA, and place it at -80°C for 40 minutes. The DNA was collected by centrifugation at 12000 rpm for 20 minutes at 4°C, washed with pre-cooled 70% ethanol, and redissolved in 50ul TE (pH8.0) after vacuum drying.
[0052] 2. Quality detection after DIG DNA probe labeling: Dilute the labeled DNA probe to the following six gradients, 1, 10 -1 、10 -2 、10 -3 、10 -4 、10 -5 . Dilute labeled control DNA to concentrations of 1 ug / ml, 100 ng / ml, 10 ng / ml, 1 ng / ml, 100 pg / ml,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com