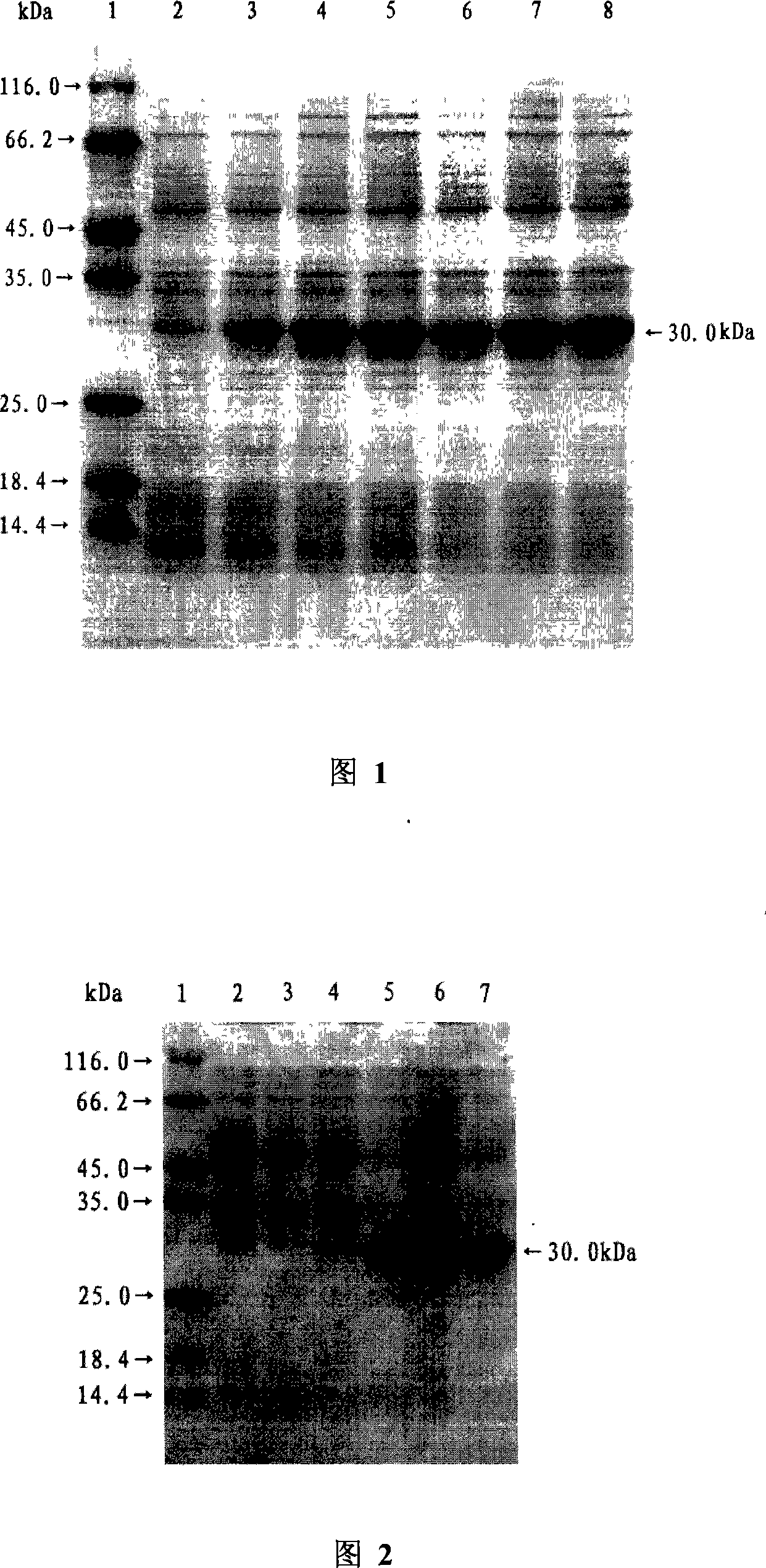
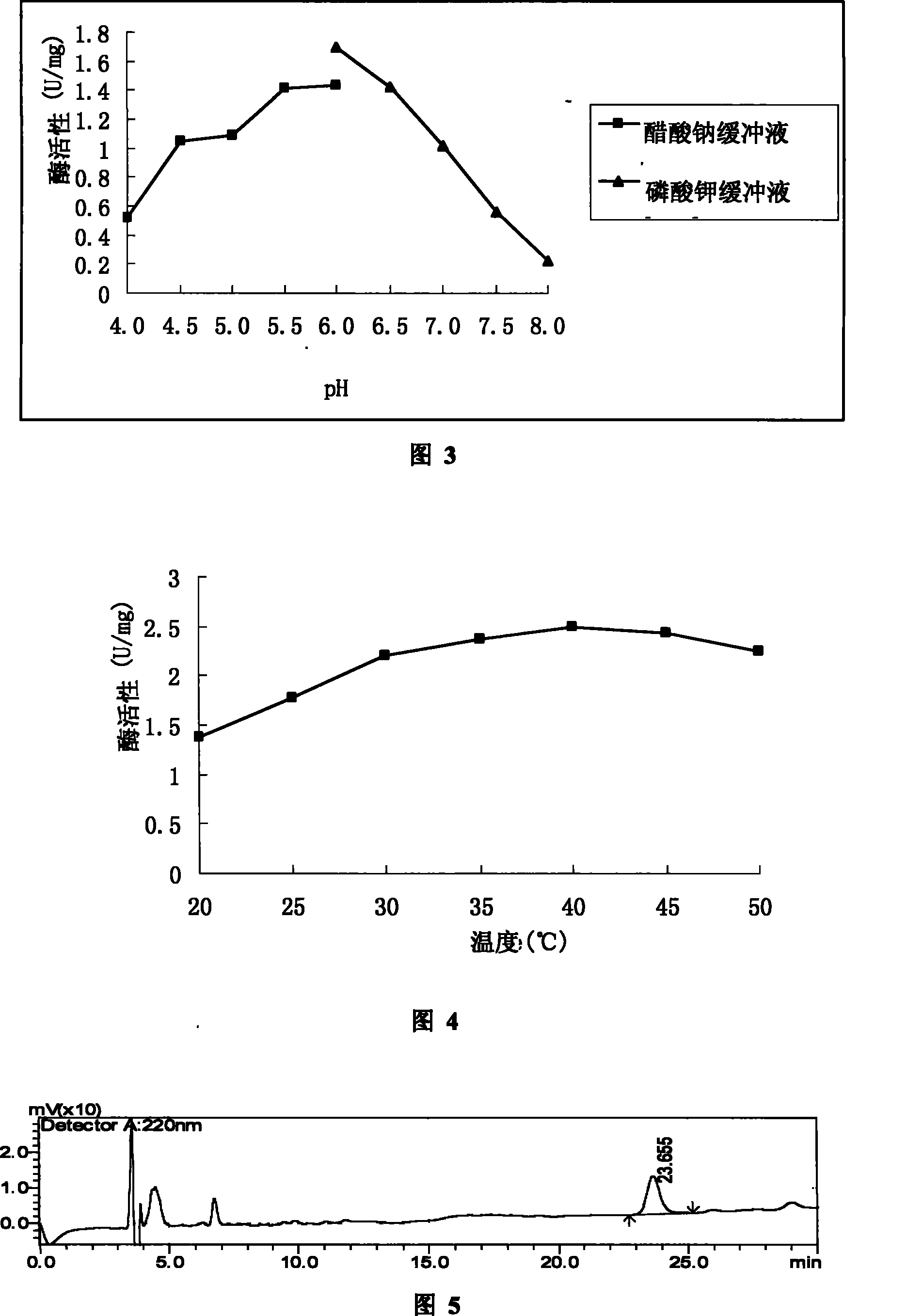
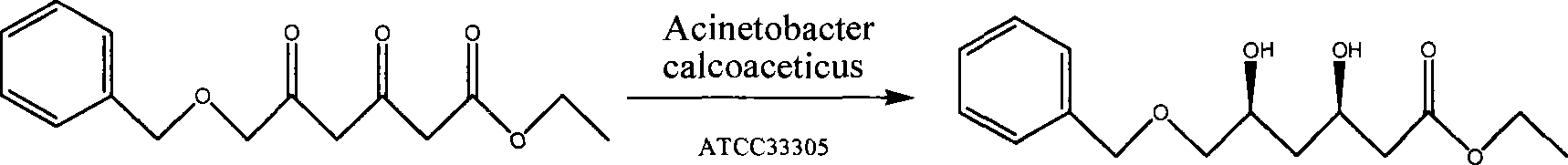Di-carbonyl reduction enzyme, its gene and uses thereof
A technology of carbonyl reductase and reductase, which is applied in the direction of oxidoreductase, application, genetic engineering, etc., can solve the problems of ignorance of complete gene and protein sequence, no direct evidence, etc., and achieve high efficiency, selectivity, and high selection sexual effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1: Cloning of Dicarbonyl Reductase Gene
[0034] 1. Bacteria and culture conditions
[0035] The strain Acinetobacter sp. ATCC33305 was purchased from ATCC in the United States and grown in the medium according to the instructions. The grown strain was stored in a glycerol cryovial at -70°C.
[0036] Glycerol cryovials were cultured in LB medium (basic components: 10g tryptone per liter, 5g yeast extract, 10g NaCl) at 37°C, 250rpm for 20 hours, then centrifuged to collect bacteria, and the method of adding lysozyme with SDS After cell disruption, genomic DNA was obtained by the phenol-chloroform method.
[0038] The amino acid sequence TGITNVTV of the N-terminal part (primer NO. 1: 5'-ACIGGIATIACIAAYGTIACIGTI-3') and the known intermediate peptide sequence GELAPAK (primer NO. 2: 5'-GGIGARCTRGCICCIGCIAAR-3') are known according to dicarbonyl reductase Design random primer sets, where "A", "G", "C", "T", "I" represent adenine, guanine, ...
Embodiment 2
[0040] Example 2 Expression of double carbonyl reductase
[0041] To facilitate the expression of the dicarbonyl reductase gene, compatible restriction sites were designed at the 5' and 3' ends of the oligonucleotide primers. Its primer pair is primer NO.3 (5'-ATACGTGC CATATG ACCGGCATCACGAATGTCACCGTTCT 3') and primer NO.4 (5'-ATACGTGCAGATACGTGCA GGATCCTCAGTACCGGTAGAAGCCCTCG-3'), with NdeI and BamHI restriction sites, respectively. Using 5.2kb KpnI DNA fragment as template, primer NO.3 and primer NO.4 as primer pair, PCR amplification was carried out. After agarose gel electrophoresis, the target gene linked to the primers was recovered with a gel recovery kit. The target gene and pET22b(+) plasmid were simultaneously digested with NdeI and BamHI, respectively, and T4 DNA ligase was used for ligation reaction. After purification, the ligated product was transformed into the competent state of E. coli BL21(DE3) strain, and coated on Incubate overnight at 37°C in LB dishes c...
Embodiment 3
[0044] Example 3 Determination of the activity of dicarbonyl reductase
[0045] The activity of dicarbonyl reductase was measured by two methods: HPLC and UV absorption. The methods and results of the measurement are as follows:
[0046] 1 HPLC method
[0047] The reaction system used in this method is:
[0048] Dicarbonyl substrate 2.5mM
[0049] NAD + 0.5mM
[0050] Formate Dehydrogenase 2U
[0052] Dicarbonyl Reductase 0.1-2mg
[0053] Make up to a final volume of 0.5 ml with 0.1 M phosphate buffer pH 6.0. The reaction was carried out at 28°C, 200 rpm for 18 h, and then stopped with an equal volume of ethanol. Mix well, centrifuge for 5 min, and use the supernatant for HPLC (Shimadzu, Japan) detection. The elution solvent is A. water containing 0.1% TFA; B. acetonitrile containing 0.1% TFA. The gradient of HPLC elution was 20-90% B with a gradient time of 12 min. The activity of its enzyme is 1 unit of the amount of enz...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com