Label-free optical detection method
a technology of optical detection and label-free technology, applied in the direction of assay labels, biochemistry apparatus and processes, instruments, etc., can solve the problems of no specific teaching or experimental evidence, no sensitive optical sensor based binding assay has been developed, and the method is time and labor intensiv
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Functionalization of Optical Sensor Surfaces for DNA Binding
[0071]The surface of a sensor-chip of the optical waveguide light spectroscope (OWLS) had been modified by vacuum sylanization or by traditional APTES-reaction in order to produce abundant —NH2+ reactive groups on the sensor-surface, for further chemical reactions as it is explained in detail below.
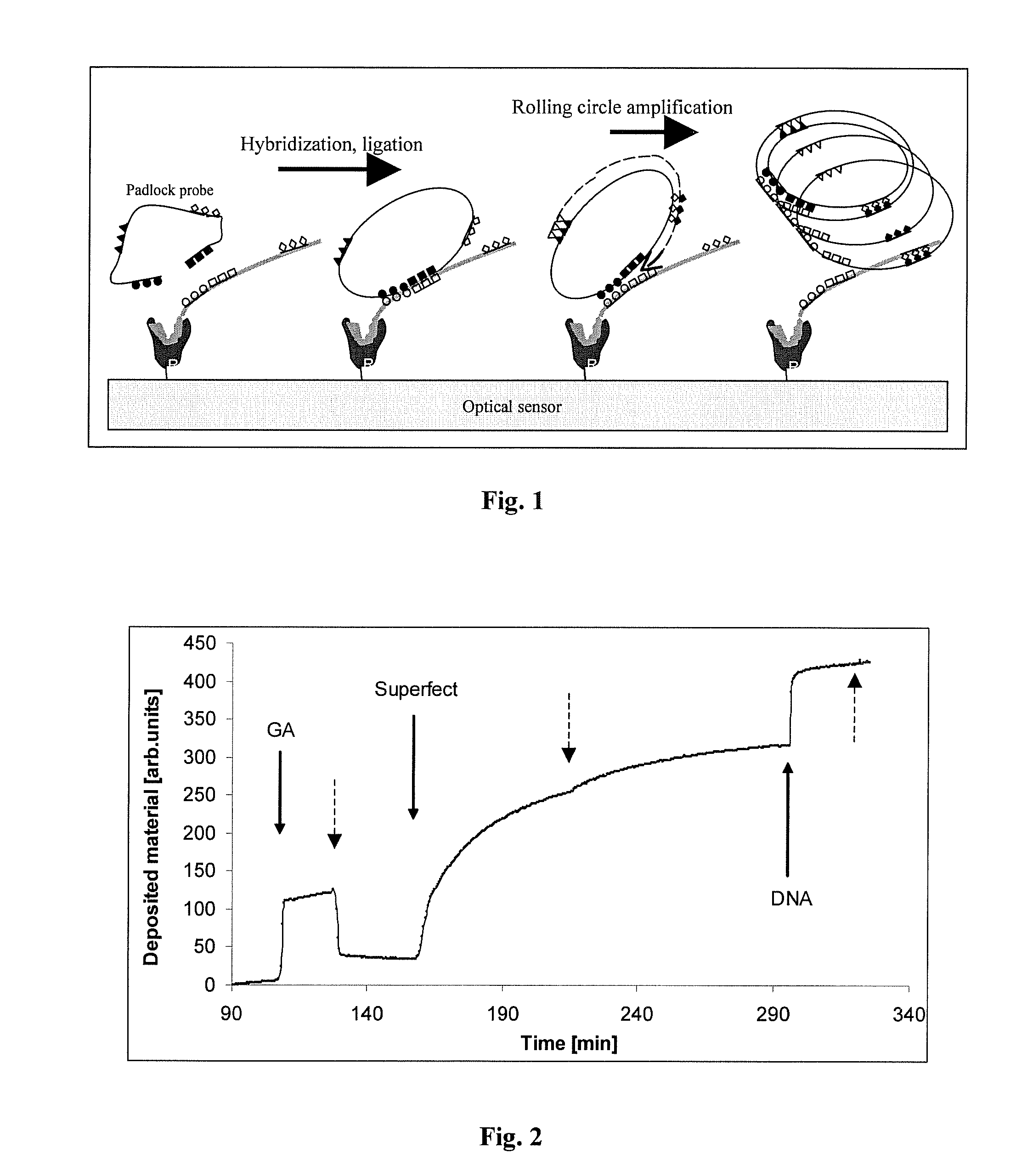
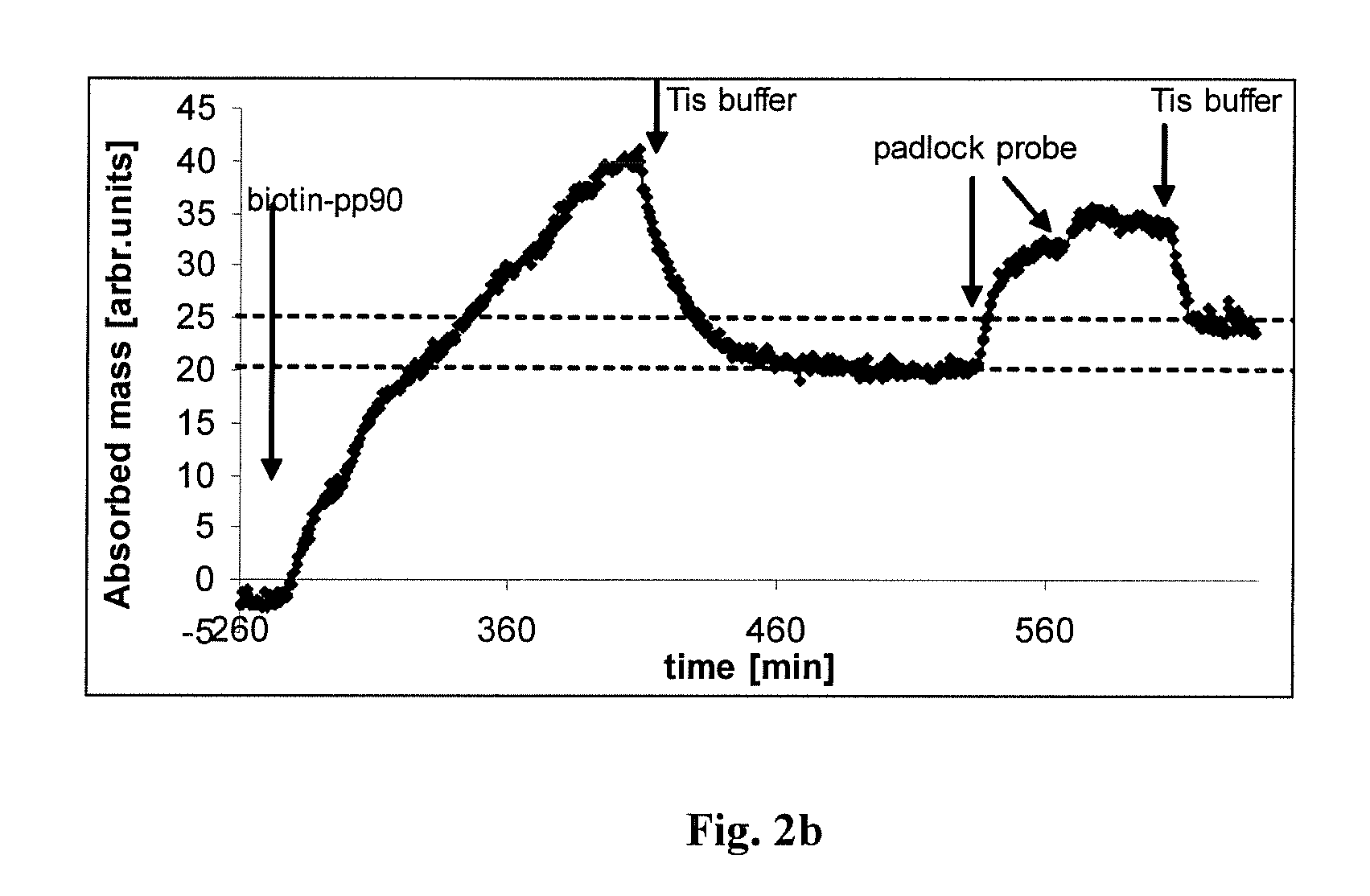
[0072]1.1. The amino-moieties were treated with glutaraldehyde, a bifunctional crosslinking molecule. The resulted glutaraldehyde-coated, reactive surface was reacted with PAMAM dendromeres resulting in a sensor-surface carrying a covalently bound lattice of DNA-binding material. (PAMAM dendromeres are materials used for introducing foreign DNA into cells, because of their strong affinity to DNA and three-dimensional particle nature, which triggers cellular endocytosis ( ).) The building of the “capturing” surface had been followed up by continuous monitoring the material deposition by real-time OWLS measurement (FIG. 2)
[0073]1.2...
example 2
Binding and Elution of DNA on Functionalized Optical Sensor Surfaces
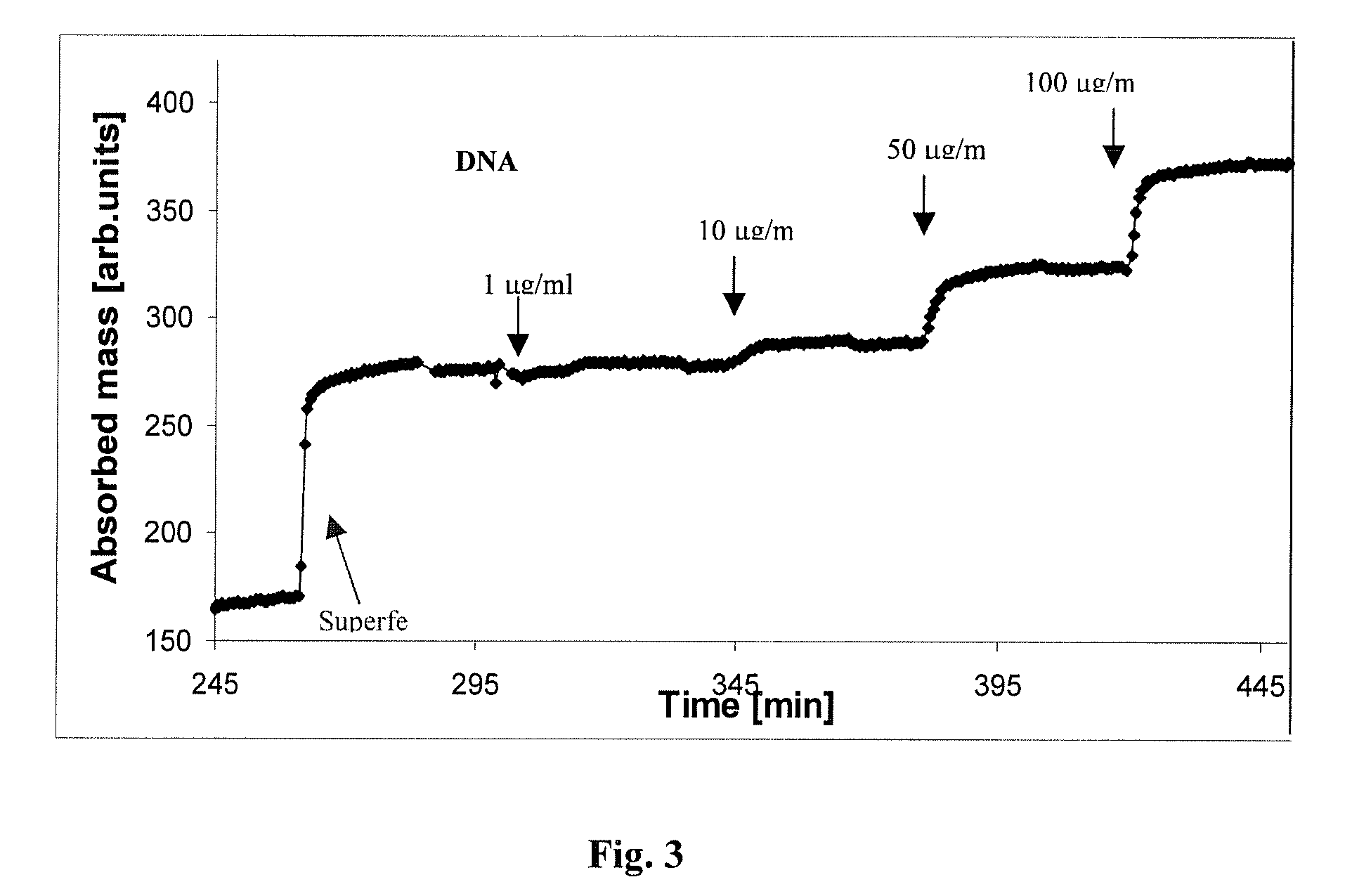
[0078]2.1. The dendromer-functionalized sensor surface captured DNA molecules from a mixture of E.Coli-derived DNA (Sigma) (last step on FIG. 2. and FIG. 3) in a concentration-dependent way (FIG. 3). The stability of DNA-binding was probed by washing with a number of different solutions including Tirs+200 mM Mg2+, SSC, 1.5 M Sodium-acetate, 0.05 M HCl, 2-10 mM EDTA in Tris buffer (TE). None of the eluting solutions could remove the captured DNA.
[0079]The binding could be broken up by washing with low ionic strength solutions at high (>70° C.) temperature (FIG. 3). The conditions for disruption of dendromer-DNA interaction suggest that the binding forces, which stabilize the DNA—dendromer complexes are similar to those coupling DNA double strands together.
[0080]High temperature washing, on the other hand, did not remove the capturing Superfect layer (FIG. 3), thus the functionalized sensors could be used repeatedly.
[...
example 3
Size-Dependent DNA Binding on Sensor Surfaces
[0082]The DNA binding was further analyzed by probing the dendromer-coated sensors with DNA fragments of different size. DNA fractions containing fragments with various lengths were prepared by sonication of the genomic E.Coli DNA (FIG. 4 A).
[0083]50 μg / ml solutions of each fraction were loaded into the OWLS cuvette and the binding was determined under identical conditions. The analyses demonstrated that relatively small DNA fragments used for specific hybridization and molecular recognition can be stably bound to the sensor surface (FIG. 4 B).
[0084]The experimental data demonstrated that relatively small fragments of DNA, those used for specific biomolecular recognition (as in situ hybridization or PCR amplification) can be stably bound to the sensor surface, and the binding can be followed up by real-time optical detection.
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| mass | aaaaa | aaaaa |
| mass change | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com