Ganglioside-associated recombinant antibodies and the use thereof in the diagnosis and treatment of tumors
a recombinant antibody and ganglioside technology, applied in the direction of fused cells, drug compositions, immunological disorders, etc., can solve the problem that the therapeutic effectiveness of gangliosides has not been demonstrated in humans
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
Obtaining of Chimeric MAb P3.
[0080] The cDNA synthesis was obtained by a reaction with reverse transcriptase enzyme, starting with RNA from the hybridoma producing Mab P3, as described previously. The sequence of the specific primers used in this reaction is shown:
[0081] For VH:
9 5' AGGTCTAGAA(CT)CTCCACACAC AGG(AG)(AG)CCAGTGGATAGAC 3'
[0082] For VK:
10 5' GCGTCTAGAACTGGATGGTGGGAAGATGG 3'
[0083] cDNA VHP3 and cDNA VKP3 were amplified by PCR using Taq Polymerase and specific primers. The restriction sites included in the primers were ECORV / NHEI, for VH and ECORV / SALI for VK. The primers sequences used were the following:
[0084] For VH:
[0085] Primer 1 (signal peptide):
11 5'GGGGATATCCACCATGG(AG)ATG(CG) AGCTG(TG)GT(CA)AT(CG)CTCTT 3'
[0086] Primer 2 (CH1):
12 5' GGGGCTAGCTGCAGAGACAGTGACCAGAGT 3'
[0087] For VK:
[0088] Primer 1 (signal peptide):
13 5' GGGGATATCCACCATGGAG(TA)CAC A(GT)(TA)CTCAGGTCTTT(GA)T 3'
[0089] Primer 2 (Ck):
14 5' AGCGTCGACTTACGTTT(TG)ATTTCCA(GA)CTT(GT)GTCCC 3'
[0090] PCR products w...
example 2
Obtaining Different Versions of the Humanized Antibody P3.
[0095] Murine VHP3 and VKP3 sequences (FIGS. 1 and 2) were compared with human sequences. FIGS. 3 and 4 show the most homologous human sequences. Helical amphipatic regions or potential T cell epitopes were searched on murine P3 variable region sequences and according with the method a judiciously strategy for aminoacid replacements was established in order to break or humanize potential T cell epitopes into the murine sequences.
[0096] The analysis on VHP3 rendered (FIG. 3) 2 amphipatic segments, the first one embraces CDR1, FR2 and some residues of the CDR2, the second one embraces the end of FR3 and CDR3. The main differences of murine sequence in comparison with the most homologous human sequence were founded in CDRs or residues involved with the three dimensional structure of the binding site. For that reason it was decided do not replace any aminoacid in murine VHP3.
[0097] The analysis for VKP3 rendered also 2 amphipatic...
example 3
Biological Activity of Chimeric MAb P3.
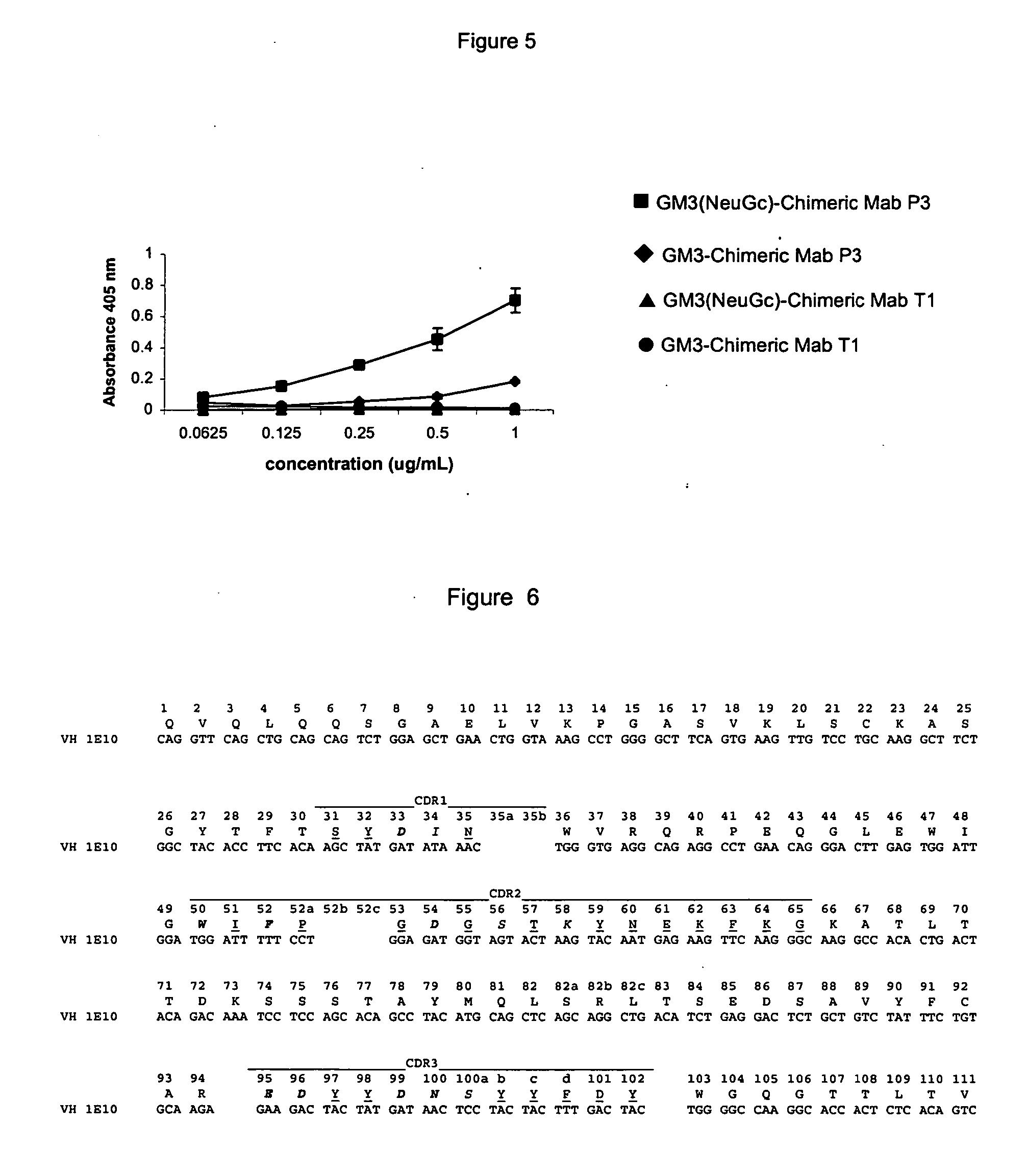
[0100] The specific binding to antigen measured by ELISA tested the biological activity of the Mab P3 chimeric.
[0101] For recombinant MAb P3, microtiter plates were coated with GM3(NeuGc) ganglioside in methanol. After drying one hour at 37.degree. C., unspecific binding was blockade with bovine sera albumin (BSA) 1% in Tris-HCl buffer, incubated for one hour at 37.degree. C. The wells were washed with PBS and incubated for 1 hour at 37.degree. C. with purified recombinant Mab P3. The wells were washed with tris-HCl and a goat anti-human antibody conjugated with alkaline phosphatase was added and incubated at 37.degree. C. for one hour. Finally, the wells were washed with Tris-HCl and the substrate buffer containing p-nitrophenylphosphate was added. After half hour absorbance at 405 nm, was measured.
[0102] Mab Ti chimeric was used as negative control.
[0103] FIG. 5 shows the specific binding of Mab P3 chimeric to the antigen.
PUM
| Property | Measurement | Unit |
|---|---|---|
| structure | aaaaa | aaaaa |
| gas chromatography- | aaaaa | aaaaa |
| affinity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com