Quick extracting method for large fragment sponge macro genome DNA
A technique of metagenomic and extraction method, which is applied in the field of rapid extraction of large fragments of sponge metagenomic DNA, can solve the problems of difficulty in extracting large fragments of DNA from sponge metagenomic, and achieves the effect of a simple method.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0014] The technical solutions of the present invention will be further described below in conjunction with the accompanying drawings and embodiments.
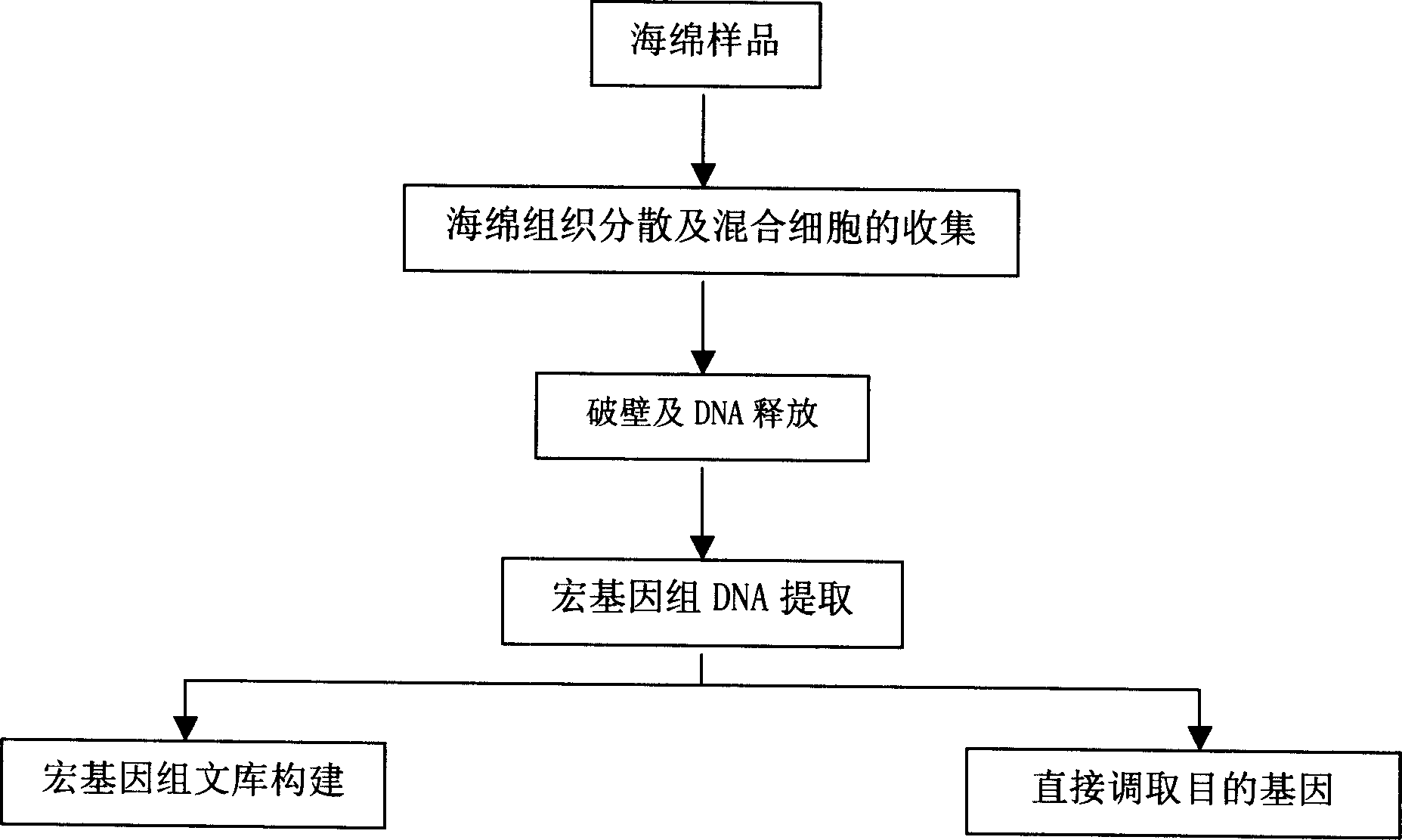
[0015] Realize the technical route of the present invention such as figure 1 Said, first obtain the sponge sample, mechanically and chemically disperse the sponge tissue to obtain mixed cells, and then digest to break the cell wall to release DNA for the extraction of metagenomic DNA.
[0016] Specific embodiments of the present invention are as follows:
[0017] 1. Spongy tissue dispersion and mixed cell collection
[0018] (1) Cut out a little spongy tissue with a sterile scalpel, put it into a 5ml centrifuge tube with an appropriate amount of sterile calcium- and magnesium-free artificial seawater (CMFSW), stir it slightly with a large-caliber pipette tip, and blow it properly;
[0019] (2) Tear into 1-3mm with sterile blunt tweezers 3 Suspend the small pieces in the original suspension and blow them with a large-caliber...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com