Recombination adenovirus construction body with deletion E1A code sequence and its use
A technology of recombinant adenovirus and construct, which is applied in the field of medical genetic engineering, can solve the problems of missing coding sequence, weak therapeutic effect of tumor cells, and low mutation rate of p53 gene, and achieve significant killing effect and the value of reliable diagnosis of tumor cells , Significant synergistic effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
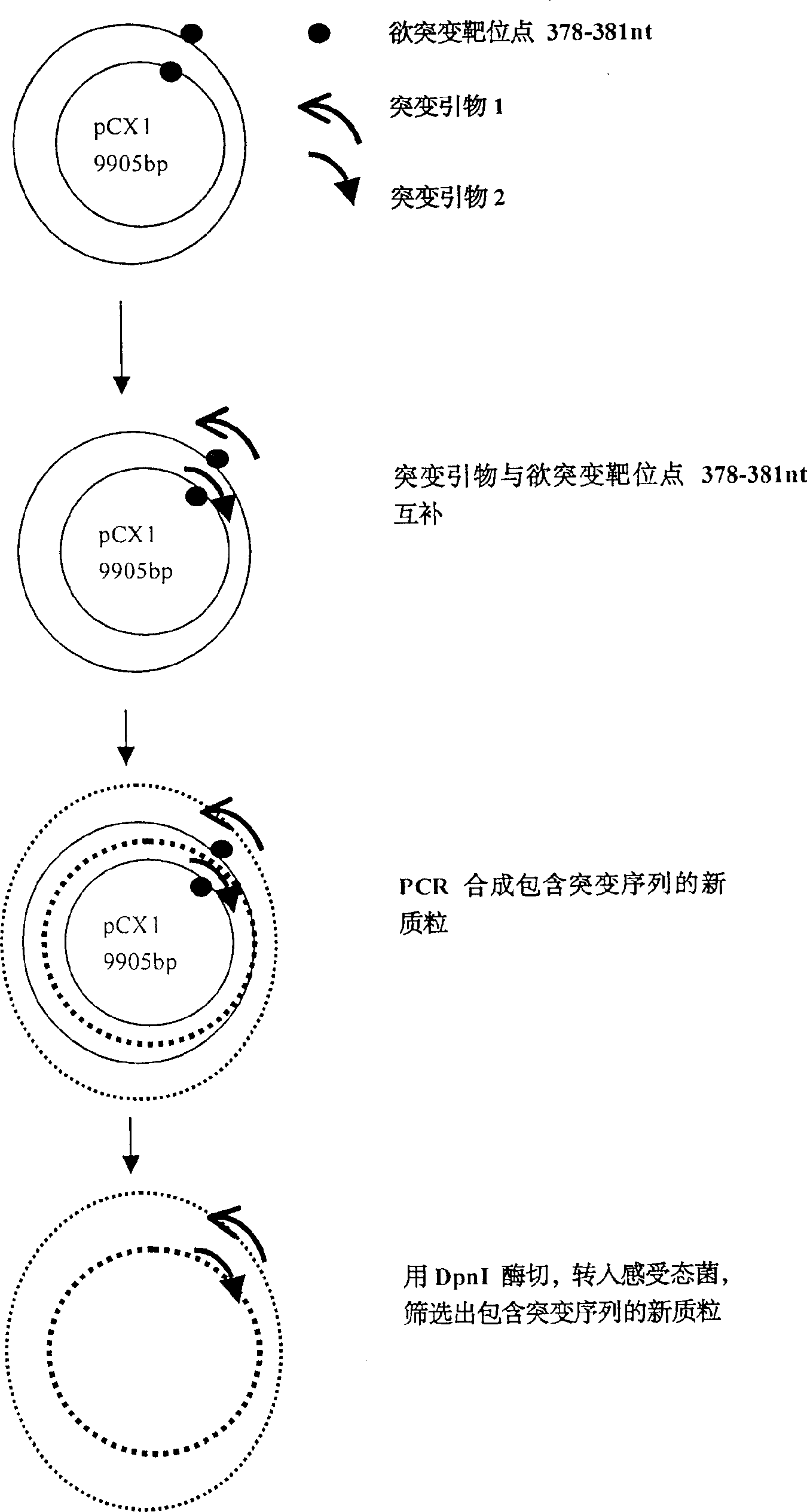
[0016] Example 1: Using site-directed mutagenesis technique to introduce Xba I restriction site in 376-381nt of pCX1 plasmid.
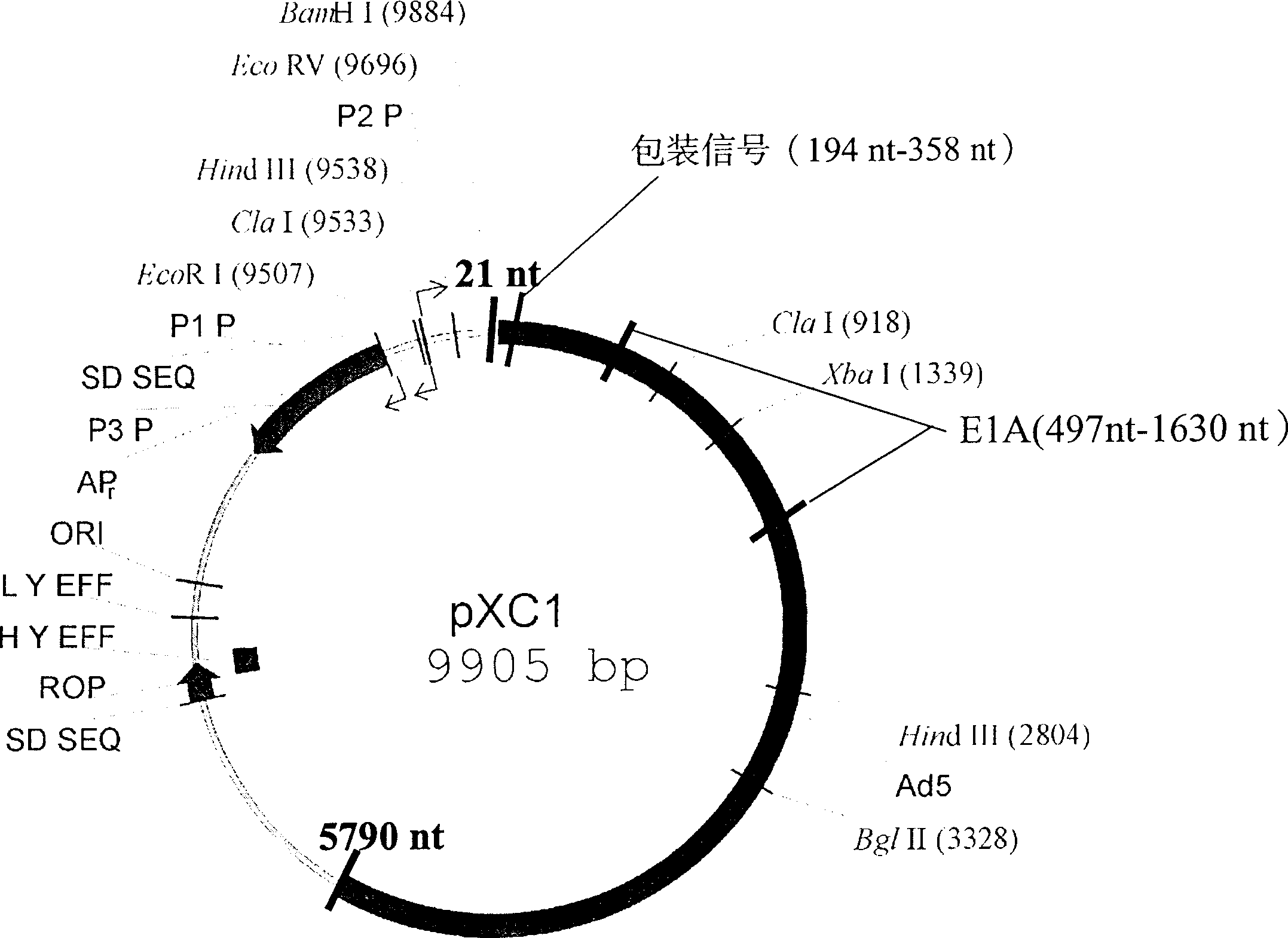
[0017] pCX1 was purchased from Microbix Biosystem Inc. (Toronato, Ontario, Canada, catalog number: PD-01-03). This plasmid contains the 21-5790 nt sequence of human adenovirus type 5, and its plasmid structure is shown in Figure 1. Site-directed mutagenesis kit QuikChange TM The Site-Directed Mutagenesis Kit (Strategene, U.S.A.) introduced the Xba I restriction site in the 376-38 1 nt of the pCX1 plasmid, that is, mutated TGG AGA to TCTAGA. The specific operation was performed according to the instructions provided by Strategene (Figure 2).
[0018] The PCR primers used for mutation are:
[0019]Primer 1 = 5'-GGG ACT TTG ACC GTT TAC GTC TAG ACTCGC CCA GGT GTT TT-3' (-358nt to -398nt);
[0020] Primer 2 = 5'-C ACC TGG GCG AGT CTA GAC GTA AAC GGTCAA AGT CCC CGC GGC-3' (-394nt to -352nt) (subscript horizontal line is the mutation site).
[0021] The ...
example 2
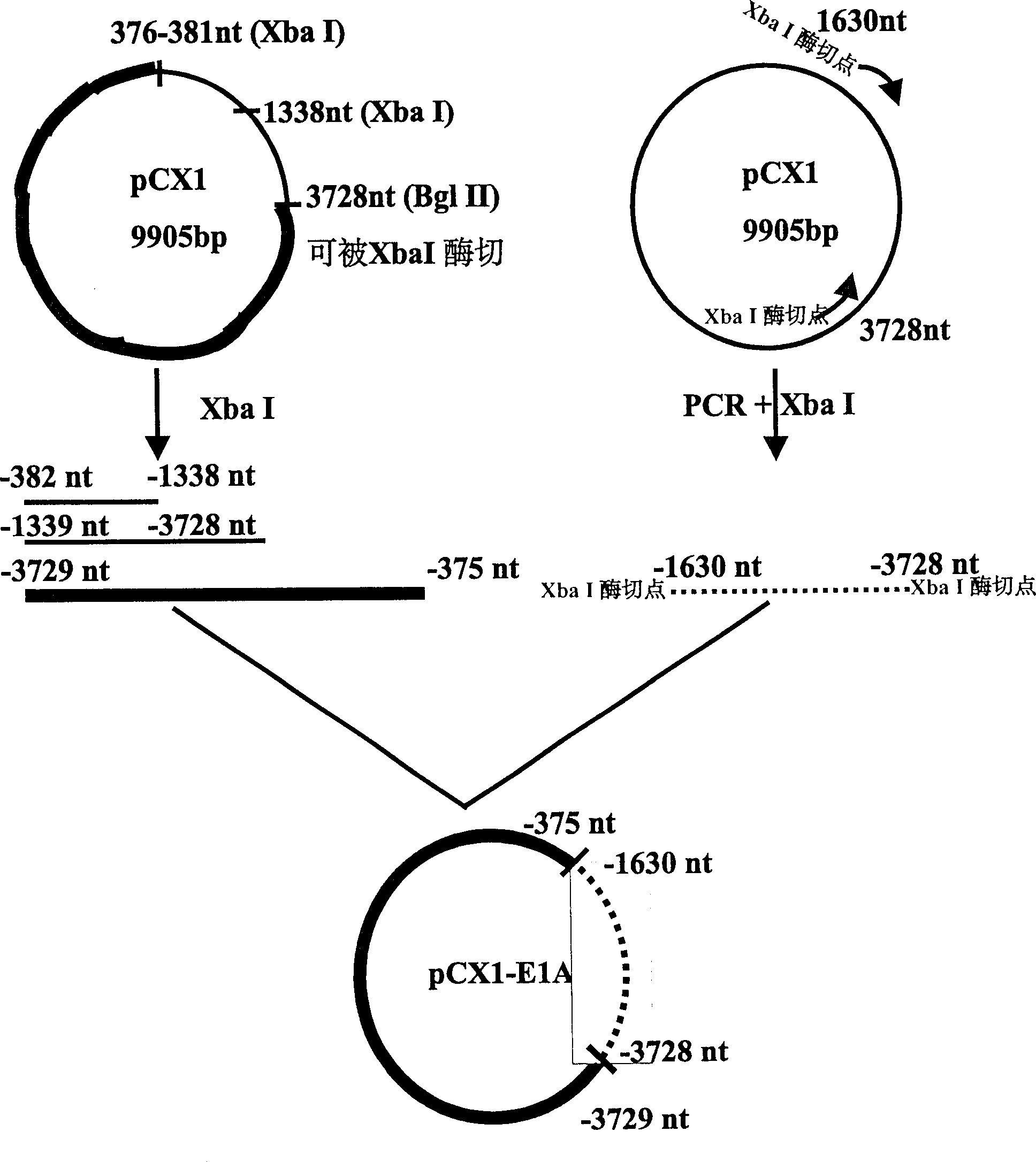
[0023] Example 2: Construction of plasmid pCX1-E1A with deletion of 382-1630 nt E1A protein coding sequence.
[0024] The PCR reaction primers are:
[0025] Primer 3 = 5'-CCG CTC TAG ATA ACT TGC ATG GCG TGTTAA-3' (-382nt to -401nt, the subscript horizontal line is the introduced enzyme cutting site XbaI);
[0026] Primer 4=5'-CCG CTC TAG ATC ATG GTC ATG TCA AACACC-3' (-1630nt to -1610nt, the subscript horizontal line is the introduced enzyme cutting site XbaI);
[0027] Using pCX1 as template, PCR reaction was carried out with primer 3 and primer 4.
[0028] The overall reaction system is 50 μl, including: 5×PCR buffer 10 μl; 50mM MgCl 2 2 μl; 10 mM dNTP 1 μl; 10 μM primer 3 1 μl; 10 μM primer 4 1 μl; pCX1 10 ng; Taq 2.5 u; add water to 50 μl.
[0029] The reaction conditions were: 95°C for 30 seconds; 25 cycles of 94°C for 30 seconds, 55°C for 1 minute, and 72°C for 1 minute; 72°C for 7 minutes.
[0030] The PCR product was a 1.7 Kb fragment, digested with Xba I, separat...
example 3
[0032] Example 3: Construction of a recombinant adenovirus construct deleting the entire coding sequence (382-1630nt) of E1A.
[0033] pBHGE3 was purchased from Microbix Biosystem Inc. (Toronato, Ontario, Canada, catalog number: PD-01-12). This plasmid contains the entire genome sequence of human type 5 adenovirus except the packaging signal (194-358nt). See the attached Figure 4. The 293 cell line is a permanent passage cell line obtained by transforming human embryonic kidney cells with cut human adenovirus type 5 (Ad5) DNA, purchased from ATCC (American typical culture collection, ATCC, U.S.A., catalog number: ATCC CRL -1573, published in 1972). At the time of acquisition, the passage number of the cells was 32 passages. 293 cells were adherent cells, low triploid, 30% of the cells had 64 chromosomes, and 4.2% of the cells had higher ploidy. Co-transfect 293 cells with pBHGE3 and pCX1-E1A, and homologous recombination occurs in the cells. When pCX1-E1A (20nt-381nt and 163...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com