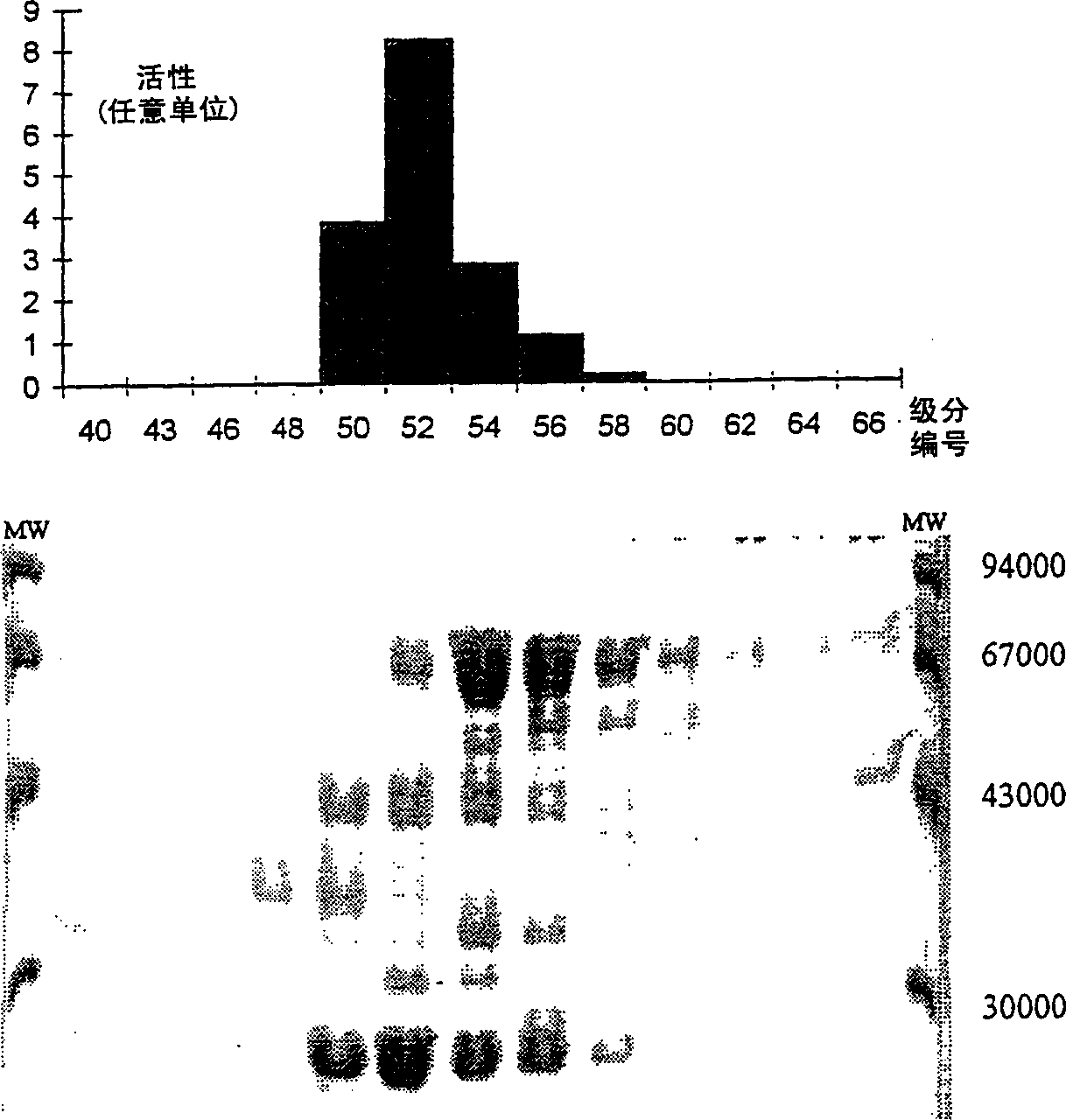
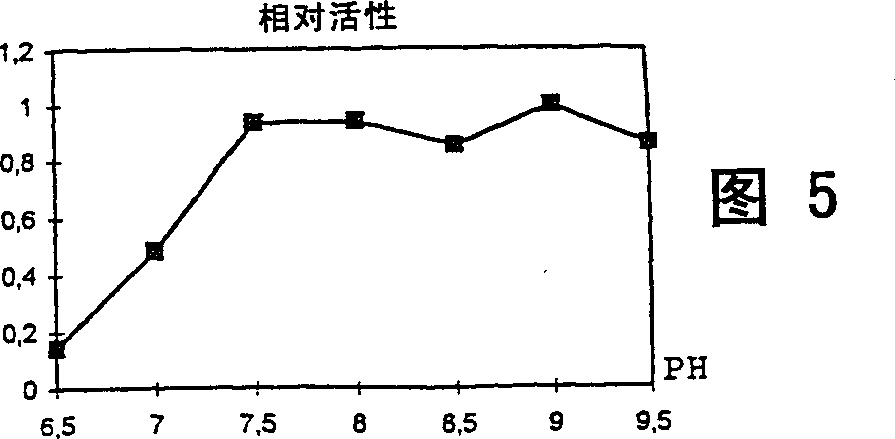
Epoxide hydrolase
An epoxide, hydrolase technology, applied in hydrolase, microorganism, microorganism-based methods, etc., can solve the problem of not describing the amino acid sequence, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] Example 1: Partial purification of epoxide hydrolase
[0070] strain
[0071] Rhodococcus rhodochrous LMGP-18079 was routinely maintained on agar slants containing 0.4% glucose, 0.4% yeast extract, 1% malt extract and 2% agar.
[0072] Determination of cis-epoxysuccinate hydrolase activity
[0073] Carry out the enzymatic reaction at 37°C in a system with a final volume of 9.3ml, which includes the following components: 0.7ml 0.1N Tris. HCl buffer (pH 8.0), 2.6ml 1.14% Triton-X100 solution, 5.0ml 30% sodium cis-epoxysuccinate solution and 1.0ml cell suspension. H 3 PO 4 The reaction was terminated by diluting the mixture 100-500 times with water acidified to pH 2.2.
[0074] L-tartaric acid formed during the reaction was determined by HPLC (flow rate between 400-600 μl / min, sample volume 20 μl) on a Vydac C18 column (Cat. No. 201HS3410) at room temperature. The solvent is the same as that used to dilute the sample. cis-epoxysuccinic acid and tartaric acid were de...
Embodiment 2
[0094] Embodiment 2: Determination of amino acid sequence of cis-epoxysuccinate hydrolase
[0095] After electrophoresis by SDS-PAGE and electroblotting on PVDF Immobilon-P membrane (Millipore), the amino-terminus of the protein was sequenced according to the general procedure. Sequencing was performed using a 477A protein automatic sequencer (Applied Biosystem) coupled to an HPLC 120A analyzer.
[0096] To determine the sequence of internal fragments, the protein was first digested with trypsin on the membrane. The resulting peptides were separated by reverse phase chromatography on HPLC, followed by amino-terminal sequencing as described above.
[0097] resulting in the following sequence:
[0098] Amino terminus: MQLNNANDNTQF SEQ ID NO 1
[0099] First internal peptide: SWPDVPSGLEQLR SEQ ID NO 2
[0100] Second internal peptide: RPLEYGPTGR SEQ ID NO 3
Embodiment 3
[0101] Example 3: Identification of cis-epoxysuccinate hydrolase gene
[0102] 1. Design oligonucleotide probes
[0103] According to the amino-terminal sequence of the protein (SEQ ID NO 1), a digoxigenine-labeled oligonucleotide was synthesized. The oligonucleotide sequence is as follows:
[0104] 5′AAYAAYGCNAAYGAYAAYAC 3′ SEQ ID NO 4
[0105] Y stands for C or T, and N stands for any one of the four bases.
[0106] 2. Hybridization of oligonucleotides with total DNA of Rhodococcus rhodochrous
[0107] 2.1 Isolation of genomic DNA
[0108] Rhodococcus rhodochrous LMGP-18079 strain was cultured overnight at 37°C in 200 ml LBroth. After centrifugation, the cells were washed at 80°C with TE buffer (Tris-HCl 10 mM, EDTA 1 mM, pH 8.0) for 30 minutes, centrifuged again, and resuspended in 15 ml of TSE buffer (Tris-HCl) containing 120 mg lysozyme. 50mM, sucrose 200mM, EDTA 1mM, pH8.0), and incubated at 37°C for 2 hours, then added 1.5ml of 250mM EDTA (pH8.0). The mixture w...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com