QRT-PCR (quantitative reverse transcription-polymerase chain reaction) method for identifying novel coronavirus Omicro variant BA-2 branch
A coronavirus, BA-2 technology, applied in biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problem of affecting the detection time, throughput and cost of virus variant strains, and the high cost of detection materials and labor , many uncontrollable influencing factors, etc., to achieve the effect of easy high-throughput operation, reducing the probability of operation pollution, and improving the detection throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] A qRT-PCR method for identifying branches of novel coronavirus Omicron variant strain BA-2, comprising the following steps:
[0045] Step 1, use the TrizoL method to extract the new coronavirus RNA;
[0046] Step 2, ARMS-based qRT-PCR primer probe design:
[0047] Step 2.1, primer design: According to the general principles of primer design, two pairs of primers are designed for each mutation site in combination with the nucleotide sequences near the mutation site of the new coronavirus Indian variant, in which the 3' ends of the upstream primers match the mutation and non-mutation sites respectively. Variation point, i.e. mutated upstream primer and non-variant upstream primer, two pairs of upstream primers share one downstream primer;
[0048] When the variant site to be identified is A27S, the nucleotide sequence of the variant upstream primer is shown in SEQ ID NO.1: 5'-CCAGAACTCAATTACCCCCTT-3', and the nucleotide sequence of the non-variant upstream primer is show...
Embodiment 2
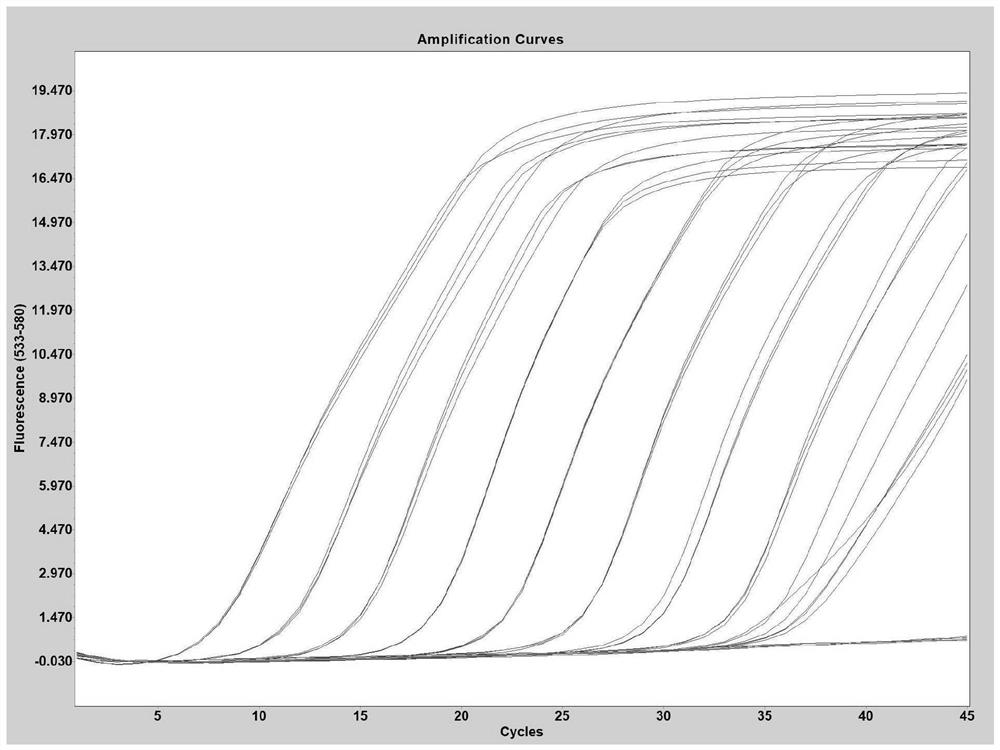
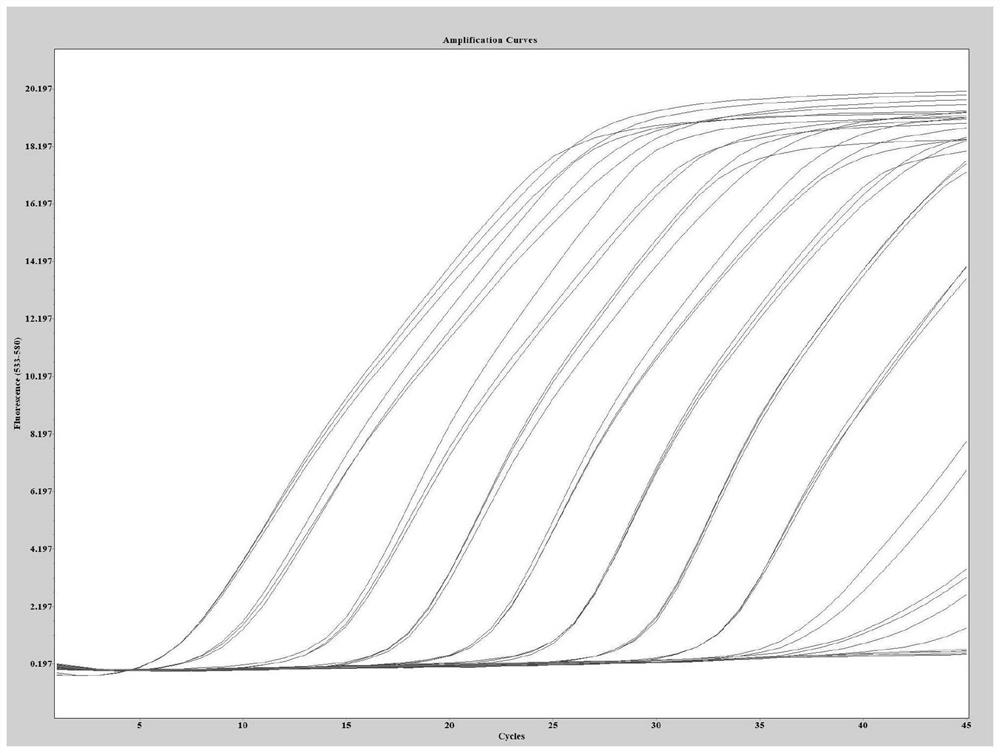
[0064] Sensitivity experiment is done to the method of the present invention (template dilution 10 6 -10 -1 copy / microliter), the experimental results are as follows:
[0065] like figure 1 As shown, when the variant site to be identified is A27S, the detection sensitivity is 7.25 copies / µl; such as figure 2 As shown, when the variant site to be identified is V213G, the detection sensitivity is 2.14 copies / µl; such as image 3 As shown, when the variant site to be identified is T376A, the detection sensitivity is 6.80 copies / µl; as Figure 4 As shown, when the variant site to be identified is R408S, the detection sensitivity is 4.21 copies / microliter.
Embodiment 3
[0067] The precision of the repeatability test evaluation method refers to the degree of closeness between a series of single measured values obtained by repeated determination of the same sample under certain conditions, and is an indicator of the size of the random error. Inter-batch repeatability (intra-day, inter-day repeatability), operator / instrument repeatability, etc.
[0068] We investigated the repeatability (precision) of this method, which was measured by the coefficient of variation. The experimental results are shown in Table 1.
[0069] Table 1 Repeatability (precision) of this method
[0070]
[0071]
[0072]Coefficient of variation (%)=(Ct standard deviation / Ct mean)×100%.
[0073] It can be seen from Table 1 that:
[0074] The intra-assay coefficient of variation of A27S is in the range of 0.46%-2.19%, and the inter-assay coefficient of variation is in the range of 0.39%-2.06%;
[0075] The intra-assay coefficient of variation of V213G is in the r...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com