Application of RNA Hsacirc0063865 inhibitor in preparation of esophageal squamous cell carcinoma resisting medicine
An inhibitor, esophageal squamous cell technology, applied in the field of biomedicine, can solve the problems of poor treatment effect and single drug treatment of esophageal squamous cell carcinoma, achieve good drug development prospects, reduce treatment costs, and inhibit proliferation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1: The expression level of Hsa_circ_0063865 in esophageal squamous cell carcinoma tissue was significantly higher than that in paracancerous tissue detected by RT-QPCR;
[0022] 1. RNA extraction from tissue samples of esophageal squamous cell carcinoma
[0023] 1) After the approval of the Medical Ethics Committee of Southeast University and the informed consent of the patients / family members, 96 patients diagnosed with esophageal squamous cell carcinoma in the First People's Hospital of Huai'an City, Jiangsu Province from 2009 to 2012 were selected, and a total of 96 cancer tissues were collected. and corresponding paracancerous tissues;
[0024] 2) Take 0.1cm 3 Cut the tissue into pieces with scissors, put it into a 1.5mL EP tube, add 1mL Trizol and 3 grinding steel balls, grind at 70Hz for 10min (the grinding box is pre-cooled in advance), and then follow the routine extraction steps of Trizol reagent;
[0025] 3) Take 1 μL of RNA sample to detect its conc...
Embodiment 2
[0042] Example 2 Hsa_circ_0063865 is involved in the malignant transformation of esophageal cancer induced by nitrosamine combined with algae toxin
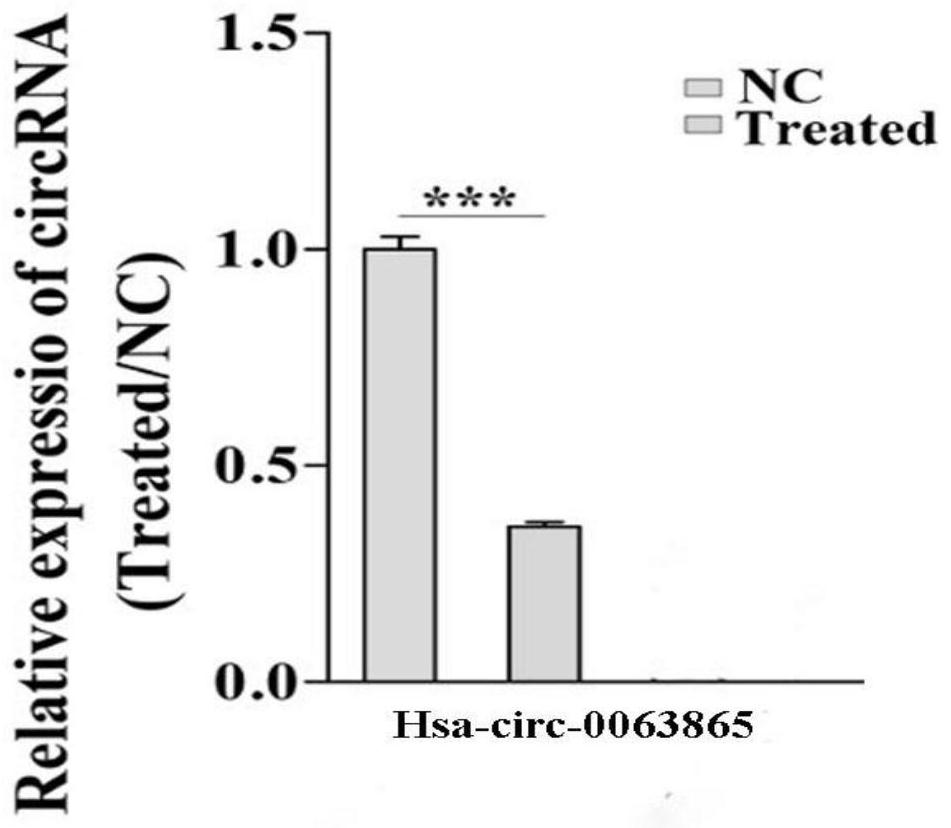
[0043]1. Construction of Hsa_circ_0063865 low-expression cell model The two Hsa_circ_0063865 siRNA sequences constructed were siRNA1: AGCGGCUCAGGCAGCUGAGG (SEQ ID NO.4) and siRNA2: CCUCAGCUGCCUGAGCCGCU (SEQ ID NO.5), and the sequence selected by NC was NC1: UUCUCCGAACGUGUCACGU (SEQ ID NO. .6) and NC2: ACGUGACACGUUCGGAGAA (SEQ ID NO. 7). The Hsa_circ_0063865 siRNA sequence was used to construct the interference lentivirus. The interference plasmid is pHBLV-U6-Puro-RFP, and the U6 promoter universal primer GGACTATCATATGCTTACCG. Subsequent steps were followed by conventional lentiviral transfection steps. The RNA of the stably transfected cell line was extracted, and the interference efficiency of Hsa_circ_0063865 was detected by RT-QPCR.
[0044] 2. Hsa_circ_0063865 is involved in the malignant transformation of esophageal cance...
Embodiment 3
[0050] Example 3 Construction of Hsa_circ_0063865-specific siRNA to verify its impact on biological phenotype
[0051] 1) Construction of Hsa_circ_0063865 low-expression stable transfection cell model: use Hsa_circ_0063865 siRNA and interference control lentivirus to transfect Het-1A-T, and use purine screening to obtain Hsa_circ_0063865 Low , Si-NC cell line.
[0052] 2) Detection of cell proliferation ability: with 1×10 4 Cells were seeded in 96-well plates per well, and EdU was incubated for 2 hours when the cells grew to 50% confluence, and the following steps were followed by BeyoClick TM According to the instructions of the EdU cell proliferation detection kit; finally, wash twice with PBS, each time for 5 minutes; discard the washed PBS, add 50 μL of new PBS to each well, observe and take pictures under a microscope; randomly select 5 fields of view in each well to take pictures, and count the green cells (proliferating cells), number of blue cells (total cells).
[...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com