Cucumber CsSTK gene, protein, expression vector and application
A technology for expressing vectors and genes, applied in the field of molecular biology, can solve the problems of long time, slow onset, no reports on cucumber STK gene related research, etc., and achieve the effects of enhanced resistance and mild symptoms.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Cloning of Example 1 Cucumber CsSTK Gene
[0040] Using the cDNA of D9320 fresh leaves 48 hours after inoculation with the pathogen Corynespora leaf spot as a cloning template, Primerpremier 6.0 was used to design specific primers for cloning the full-length CsSTK gene and perform PCR amplification.
[0041] Specific primers are as follows:
[0042] CsSTK-F: ATGTCAAAGGTTCTTGCAGC
[0043] CsSTK-R: TTACTTTGTGGTTTTGAACAACTG
[0044] 20 μL amplification reaction system, including the following components: 10 μL 2×Taq PCR MasterMix, 0.5 μL upstream primer, 0.5 μL downstream primer, 1 μL cDNA template, ddH 2 O 8 μL.
[0045] PCR amplification program: 94°C for 2min; 94°C for 30s, 52°C for 30s, 72°C for 1min, 30 cycles; 72°C for 10min; 4°C Hold.
[0046] The amplified target fragment was ligated with the pEASY-T3 vector to obtain pEASY-T3-CsSTK E. coli bacterial liquid, which was identified by PCR and sent to Harbin Qingke Biological Company for sequencing. The electrophor...
Embodiment 2
[0047] Construction and transformation of embodiment 2 plant overexpression vector
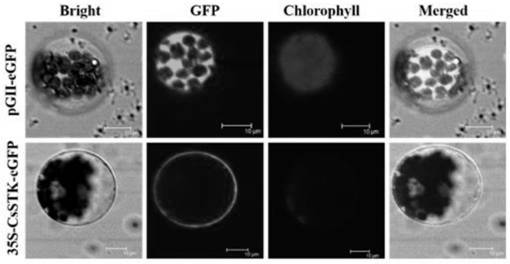
[0048] The full-length primers of the cloned gene CsSTK were used for cloning and gel recovery. The pCXSN-1250 vector is a plant expression vector, which contains a 35s promoter, and the pCXSN-1250 empty vector is single-digested with restriction endonuclease XcmⅠ. Mix the cut empty vector and the target fragment according to the ratio, and use T4 ligase to ligate at 16°C for 16 hours to obtain the pCXSN-CsSTK overexpression vector, and transfer it into DH-5α competent cells by freeze-thaw method. The culture was screened on the medium containing Kan, and colony PCR identification was carried out after 12 hours.
[0049] pCXSN-CsSTK sequencing primers:
[0050] pCXSN-1250-F: CGGCAACAGGATTCAATCTTA;
[0051] pCXSN-1250-R: CAAGCATTCTACTTCTATTGCAGC.
[0052] The amplification system and reaction procedure are the same as in Example 1.
[0053] The results of electrophoresis of the PCR product...
Embodiment 3
[0055] Embodiment 3 Transgenic positive plants are obtained
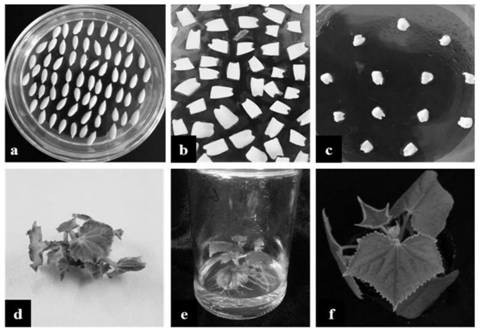
[0056] Soak the seeds in warm water for 30 minutes, remove the seed coat, sterilize with 75% ethanol for 1 minute and remove the surface tension of the seeds, then sterilize with 2-3% sodium hypochlorite for 10 minutes, rinse with sterilized water several times, remove the disinfectant on the surface of the seeds, After drying the water with sterilized filter paper, spread it evenly in the germination medium (MS solid powder 4.33g L -1 +30g·L -1 Sucrose+2g·L -1 Phytogel, pH=5.8), cultured in the dark at 28°C for 48h.
[0057] Use 1 / 2MS medium (1 / 2MS solid powder 4.33g L -1 +30g·L -1 Sucrose, pH=5.8) resuspend the above pCXSN-CsSTK overexpression vector into Agrobacterium tumefaciens cells, cut off the protective film on the surface of the above cultured seeds under sterile conditions, peel off the cotyledon, remove the growth point and hypoembryon axis, and use the bacterial liquid to infect by shaking, after t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com