A specific short peptide targeting glioma cells, its coding gene and its application
A technology for brain glioma and coding genes, which is applied in specific short peptides, coding genes and their applications, to achieve the effects of small molecular weight, good tissue penetration, and good in vivo targeting properties
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] In vitro screening of phage display random heptapeptide library and sequencing and analysis of phage monoclonal DNA
[0032] (1) Experimental process of phage display heptapeptide library screening targeting glioma cell U251
[0033] The cultured glioma cell line U251 cells were taken out and cultured in serum-free medium for 2 hours. Before the end of the incubation, 10 μL (2×10 11 pfu) was diluted into 990 μL PBS+-BSA to prepare a phage peptide library solution, which was then added to the cells and incubated in a cell culture incubator for 2 hours. Remove the phages that are not bound to the cells or non-specifically, digest the cells completely, add lysate and sterilized glycerol to store at -20°C. Take 10 μL for phage titer determination, and take half of the eluate for amplification. Increasing the washing intensity round by round is beneficial to the elution of non-specific binding phages and retaining phages with strong binding force. The amount of phage rec...
Embodiment 2
[0039] In vitro targeted identification of phage monoclonals
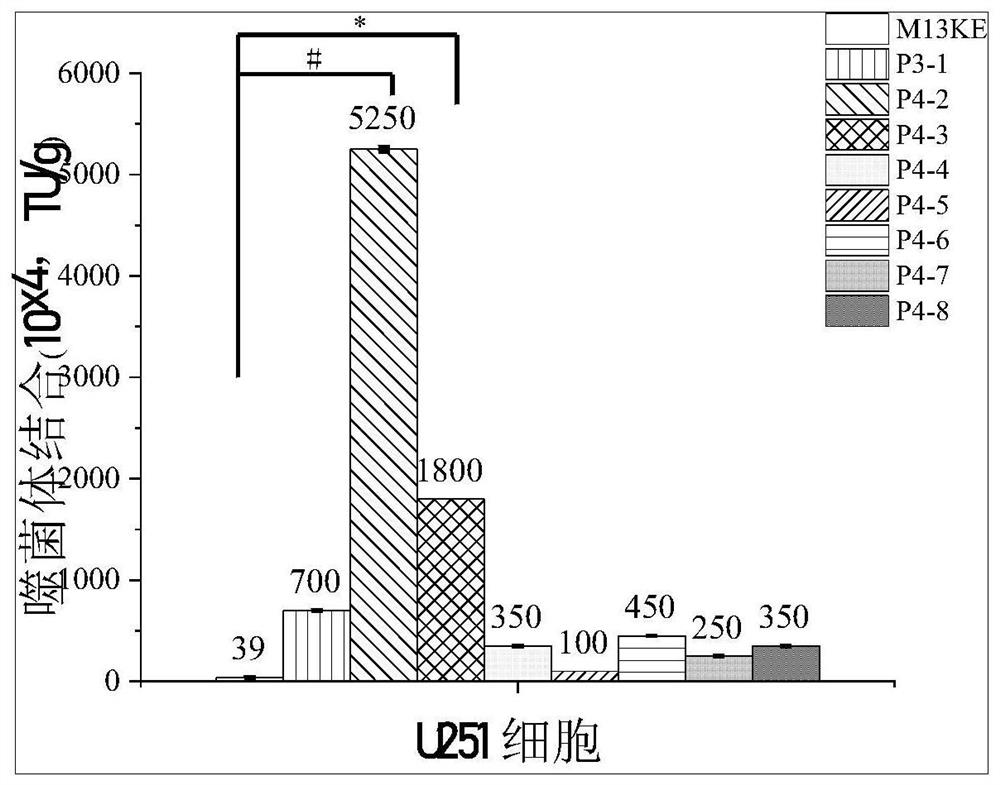
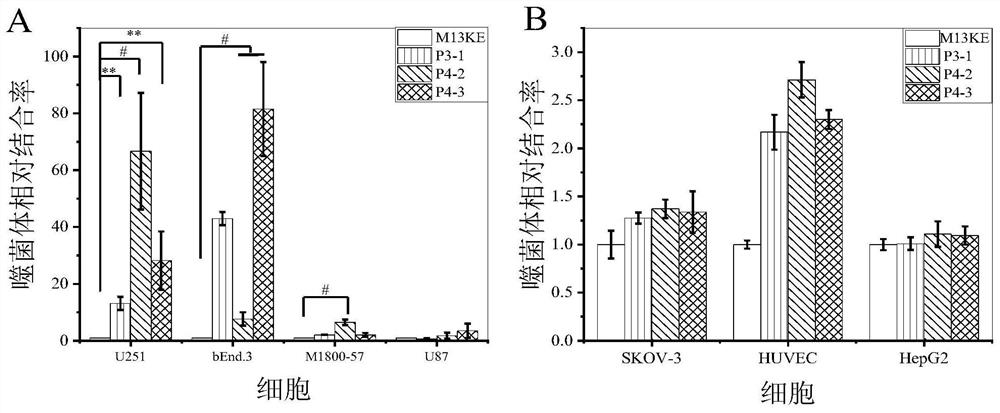
[0040] (1) Enzyme-linked immunosorbent assay (ELISA) to identify the affinity of phage monoclonal to different cells
[0041] Cells were seeded in 96-well plates and incubated in serum-free medium at 37°C. Wash twice with PBS, then add 4% paraformaldehyde, fix at room temperature, wash three times with PBS, add 3% BSA-PBS, and seal the cells at 4°C. Dilute the phage to 10 10 TU, add diluted phage to the cells, incubate at 37°C, set 4 parallels in each group, and perform 2-3 repeated experiments, with the M13 blank phage group as the control group. Wash unbound phage with PBST, add HRP / M13 phage monoclonal antibody complex diluted with 3% BSA-PBS after washing, and incubate at 37°C. Wash twice with PBS, add ready-to-use TMB chromogenic solution to each well, and incubate at room temperature in the dark, and the color will appear to the expected depth. Join H 2 SO 4 The solution terminated the reaction, and the...
Embodiment 3
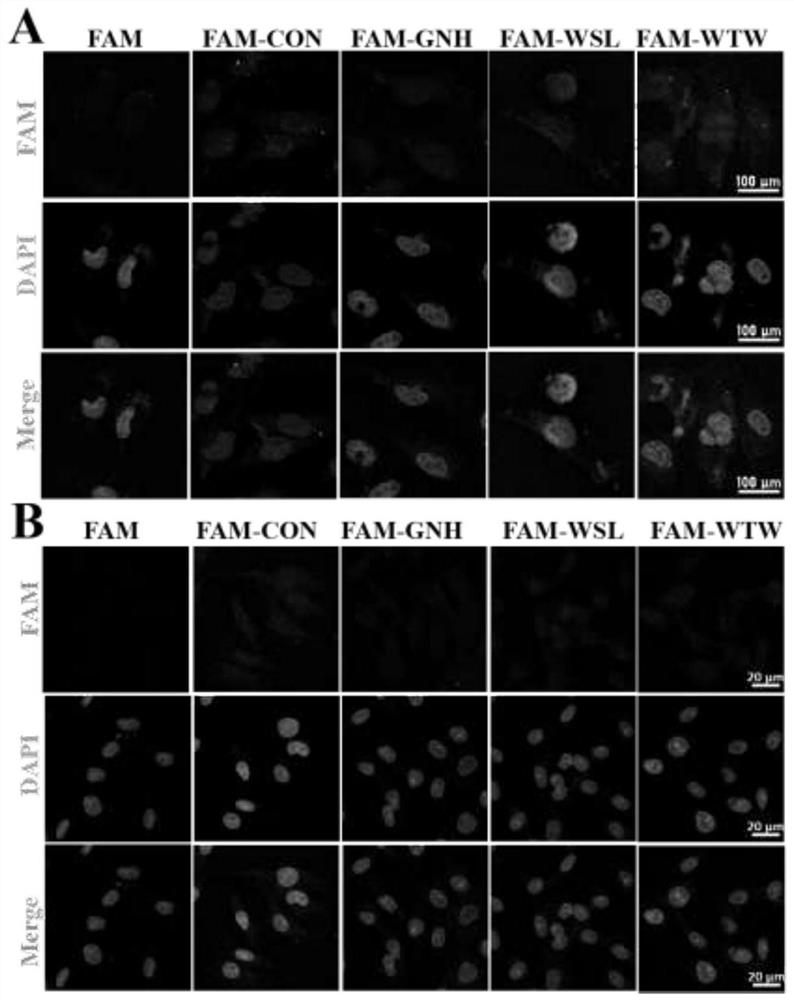
[0048] Targeted binding ability of short peptides in vitro
[0049] Three candidate short peptides and random short peptides were synthesized by Shanghai Botai Biotechnology Co., Ltd., and FAM fluorescent groups were modified at the N-terminus of the short peptides for immunofluorescence and flow cytometry experiments. U251 cells 10 4 Seed in a glass-bottom dish, and after the cells grow completely attached to the wall, they can be used for in vitro targeting identification experiments. Incubate the cells in the glass-bottom dish in serum-free medium, then add 3% BSA+PBS to block, add 0.1% Triton to permeabilize, and incubate with fluorescent short peptide in the dark for 1 hour, then add DAPI to counterstain the nucleus , observed on a confocal laser microscope.
[0050] The experimental process of flow cytometry also requires seeding U251 cells on a 6-well plate, culturing them in serum-free medium and then incubating them with fluorescent short peptides, digesting the cel...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com