MYB transcription inhibition factor LrETC1 related to lycium ruthenicum anthocyanin synthesis and application thereof
A technology of transcriptional repressor and qianthocyanidin, which is applied in the field of genetic engineering and can solve problems that have not been reported
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] For the extraction of DNA and RNA, refer to the kit instructions, and operate in a conventional manner.
[0063] Synthesis of single-stranded cDNA: Using the total RNA of Lycium barbarum fruit as a template, the single-stranded cDNA was synthesized using the experimental steps given by Takara's PrimeScriptTMRT reagent Kit with gDNA Eraser (Perfect Real Time).
[0064] Cloning of LrETC1 gene: See Table 2 for the sequences of the amplification primers. The primers were diluted to 10 μM, and the obtained cDNA was used as a template for PCR amplification reaction. The reaction system is shown in Table 3 below.
[0065] Table 2
[0066]
[0067] table 3
[0068]
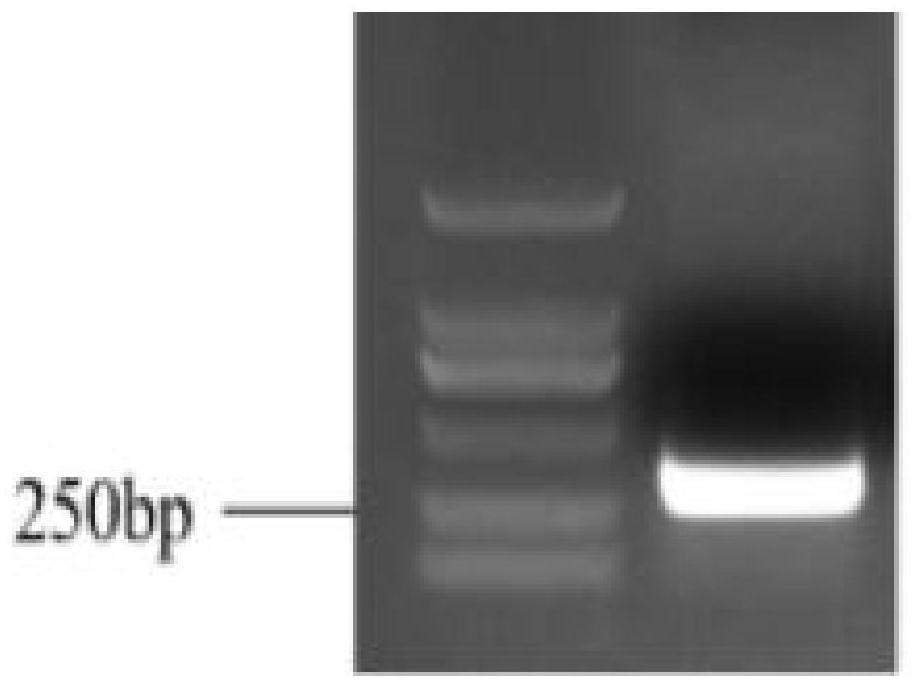
[0069] After the amplification is completed, take out the PCR product and perform 1% agarose gel electrophoresis detection. After electrophoresis for about 7 minutes, observe it under a UV gel imager. If the target band is single and the size is correct, cut out the gel piece and place it in the In a 2mL ce...
Embodiment 2
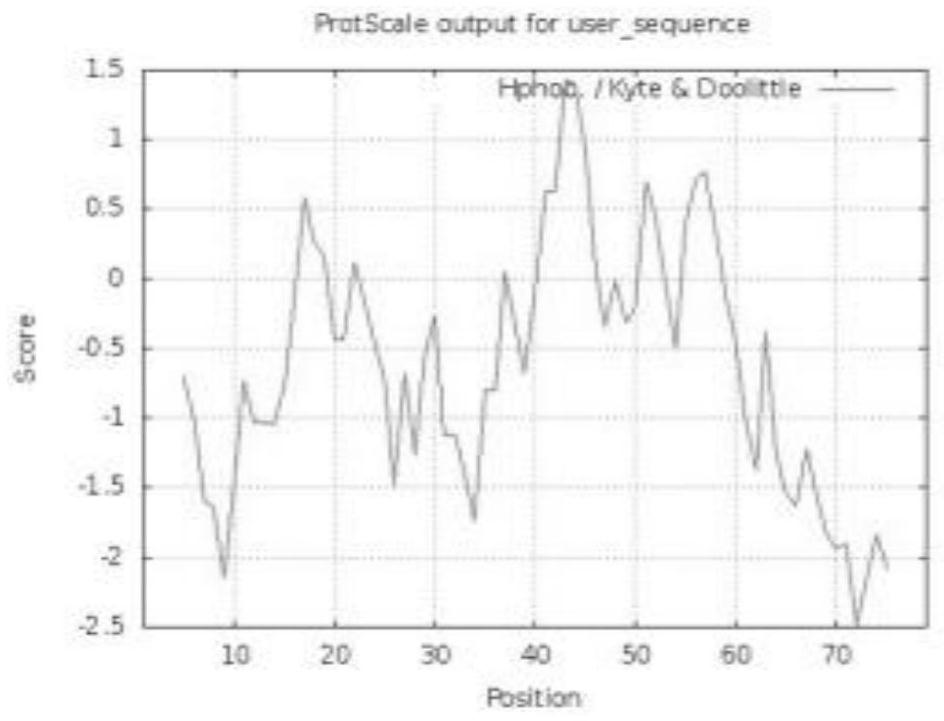
[0086] The open reading frame and deduced amino acid sequence of the LrETC1 transcription factor were searched through the ORF Finder (https: / / www.ncbi.nlm.nih.gov / orffinder / ) online search tool on NCBI. The online software ProtParam (http: / / web.expasy.org / protparam / ) was used to predict the amino acid composition, protein molecular weight, theoretical isoelectric point and stability of transcription factors. The hydrophobicity and charge distribution of proteins were analyzed using the online software ProtScale (https: / / web.expasy.org / protscale / ). The transmembrane domain of LrETC1 protein was analyzed using TMHMM 2.0 (http: / / www.cbs.dtu.dk / services / TMHMM) software. The signal peptide of the protein was predicted by Cell-PLoc2.0 (http: / / www.cbs.dtu.dk / services / SignalP). Use Wolf Psort (https: / / www.genscript.com / wolf-psort.htmL) and online software (https: / / www.csbio.sjtu.edu.cn / bioinf / Cell-PLoc-2 / ) to predict proteins subcellular localization. Protein analysis using SOPMA ...
Embodiment 3
[0163] Construction of Plant Overexpression Recombinant Vector
[0164] Cloning of target gene homologous recombination fragment:
[0165] Using the cDNA solution obtained in the steps of the above examples as a template, the corresponding primers in Table 12 were used to amplify the homologous recombination fragment of the plant overexpression vector. The amplification system, detection and recovery of PCR products are the same as those in the above examples.
[0166] Table 12
[0167]
[0168]
[0169] Vector digestion reaction
[0170] The vector pCM1307 was double-digested with two restriction enzymes XbaI and KpnI; the vector pZYB9-pEAQ-HT was double-digested with two restriction enzymes SmaI and StuI. The enzyme digestion system and product recovery are the same as those in the above-mentioned examples.
[0171] Homologous recombination connection fragments and vectors, Escherichia coli DH5α transformation, bacterial liquid PCR identification and plasmid extract...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Theoretical molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com