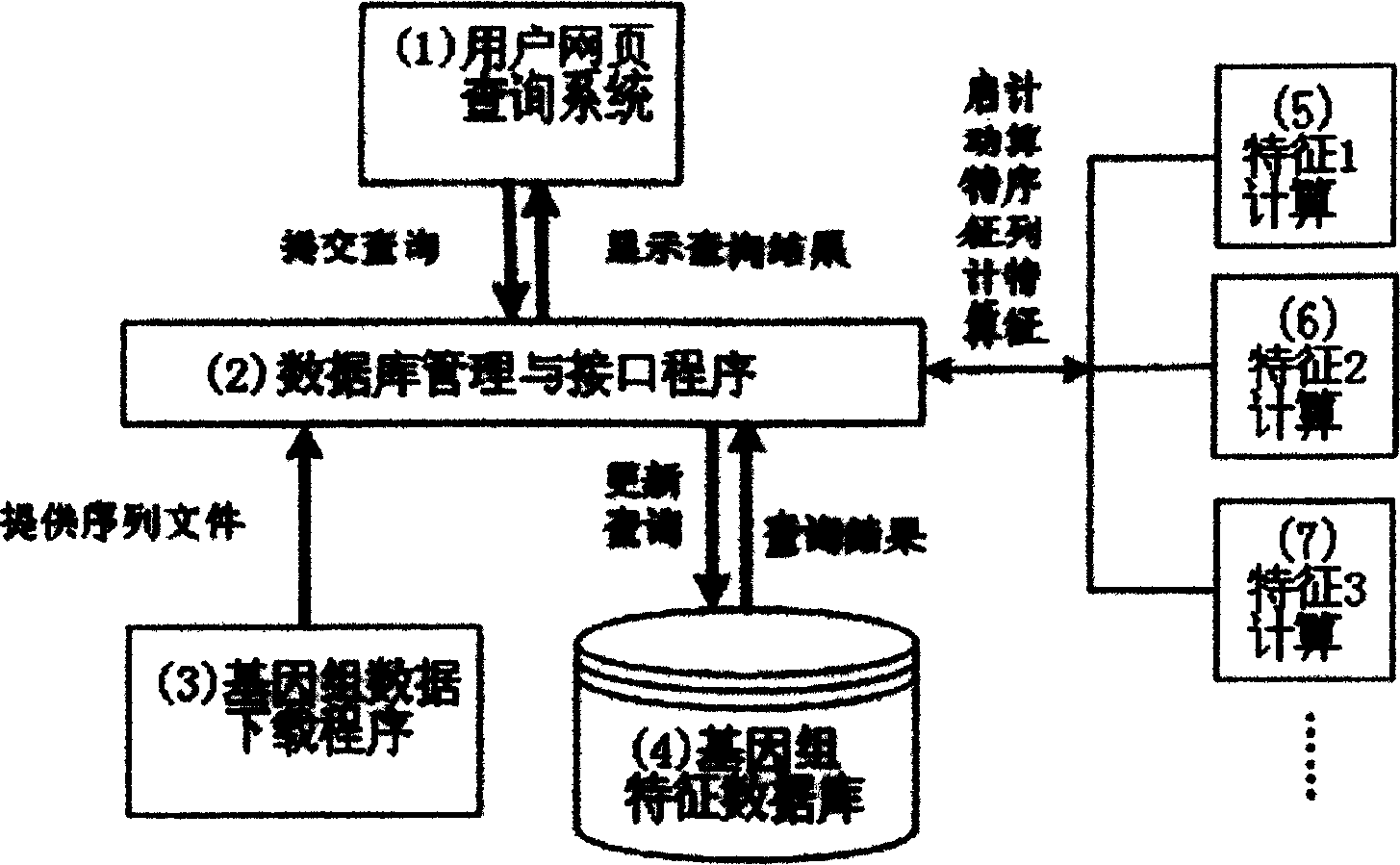
Seaching method of genome sequence data based on characteristic
A genome sequence and search method technology, which is applied in the search field of a feature-based genome sequence database, can solve problems such as missing sequences and inability to handle long sequence fragments, and achieve the effect of improving efficiency and scalability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1: Search for 100 sequences in the database that are similar in base correlation characteristics to a certain sequence.
[0045] On human chromosome 7, a sequence is selected, and its base correlation feature value is searched in the database to obtain 100 most similar sequences, such as Figure 3 ~ Figure 7 As shown, most of the 100 sequences were found to be from the human genome, and only from the 73rd sequence did some mouse genome sequences appear, indicating that the sequences in the human genome still have the characteristic of base correlation. A fairly high degree of similarity, and some fragments of the mouse genome also have a certain similarity with the human genome.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com