L-aspartic acid-alpha-decarboxylase mutant and application thereof
A technology of aspartic acid and decarboxylase, applied in the field of genetic engineering, can solve the problem of less PanD, etc., and achieve the effect of increasing the level of self-shearing and increasing the yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
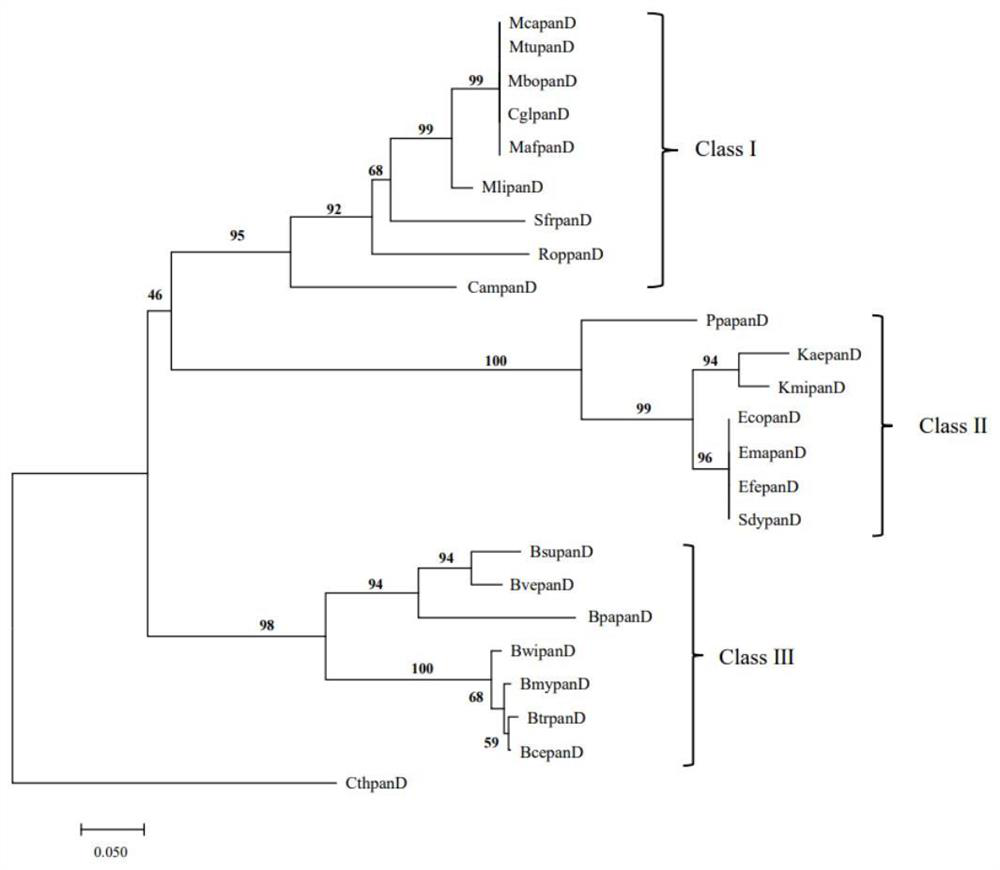
[0053] Example 1: analysis of panD gene molecular evolution relationship and selection pressure analysis
[0054] (1) Analysis of molecular evolution relationship of panD gene
[0055] In this study, we explored the sequence differentiation of the panD gene in different organisms through the method of molecular evolution. Using the Escherichia coli gene EcopanD (SAMN02604091) as the original template, a comprehensive search was performed on the sequence of the panD gene family through the NCBI website (https: / / www.ncbi.nlm.nih.gov / ). The confirmation standard is that the sequences with E value 40% are members of the panD gene family. Finally, the PanD amino acid sequences of 24 microorganisms were obtained, namely Mycobacterium tuberculosis (MtupanD), Mycobacterium africanum (MafpanD), Mycobacterium canettii (McapanD), Mycobacterium bovis (MbopanD), Bacillus cereus (BcepanD), Mycobacterium liflandii (MlipanD), Streptomycesfradiae ( SfrpanD), Rhodococcus opacus(RoppanD), Cory...
Embodiment 2
[0063] Example 2: Construction of L-aspartic acid-α-decarboxylase mutants by site-directed mutagenesis of the whole plasmid
[0064] (1) Primer
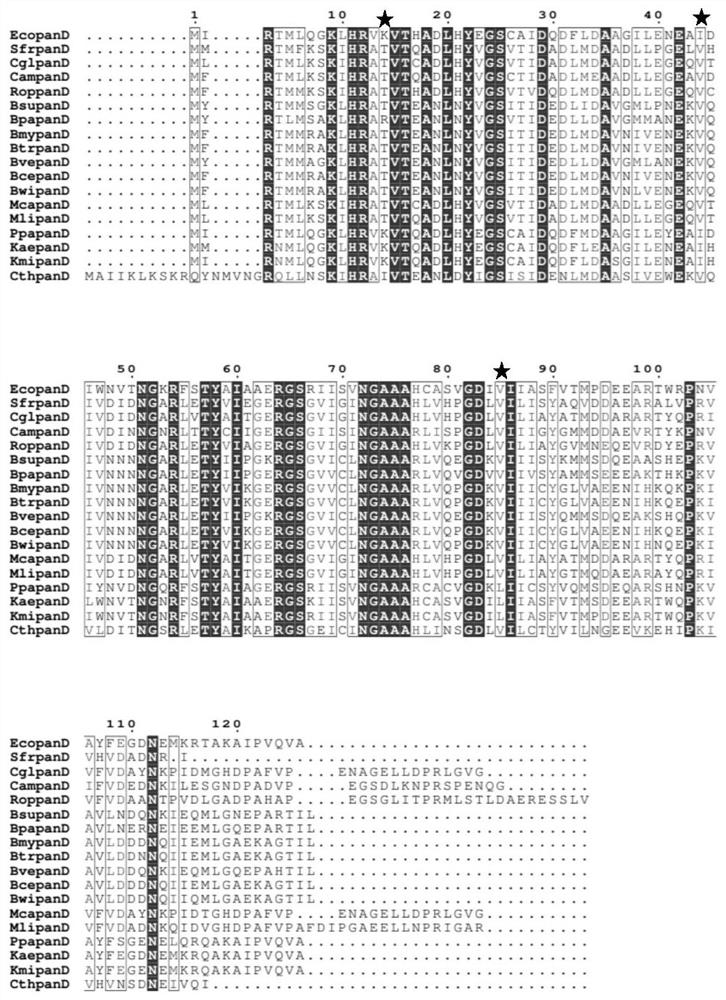
[0065] Further, on the basis of the EcoPanD protein and the enantiosite in Class III as a control, directed evolution was carried out on the above three sites to form a library of L-aspartic acid-α-decarboxylase EcoPanD mutants, which are as follows: (1) Lysine at the 14th amino acid position shown in SEQ ID NO.1 is mutated into threonine (EcoPanD K14T , the nucleotide sequence of the mutant is shown in SEQID NO.2); (2) the amino acid 44th isoleucine shown in SEQ ID NO.1 is mutated into valine (EcoPanD I44V , the nucleotide sequence of the mutant is shown in SEQ ID NO.3); (3) the amino acid 85th valine shown in SEQ ID NO.1 is mutated into leucine (EcoPanD V85L , the nucleotide sequence of the mutant is shown in SEQ ID NO.4). The PCR primer sequences used in this embodiment are listed in Table 2. In vitro amplification of the Ecopa...
Embodiment 3
[0073] Example 3: Screening and Fermentation of Mutant Library
[0074] The transformant that obtains of embodiment 2 is carried out as follows:
[0075] 1. Sequencing verification: use a sterilized pipette tip to select 3 transformants on the plate and inoculate them in LB liquid medium containing 100 μg / mL kanamycin. 100μg / mL Kana antibiotics in LB liquid medium. 37°C, shake culture at 200r / min for 12h, sequence and preserve bacteria. The constructed strains are shown in Table 3.
[0076] The strains constructed in Table 3 Example 3
[0077]
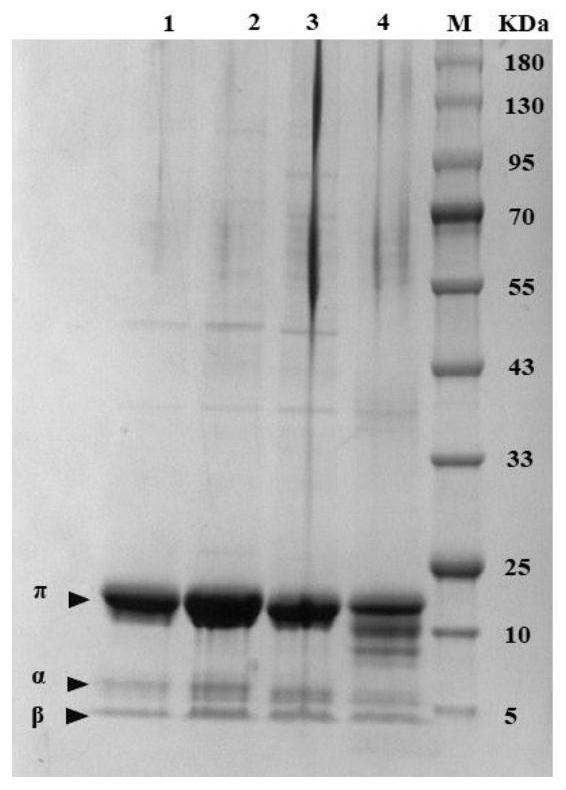
[0078] 2. Induced expression of mutants: Under sterile conditions, take 1 mL of seed liquid from the LB liquid medium in step 1, transfer to 50 mL of LB medium (final concentration is 100 μg / mL Kana), 37 ° C, 200 r / mL Shake culture for about 2 hours until OD 600 0.6 to 0.8, the final concentration of 0.3mM IPTG was added and shake cultured for 12h for expression. Crude protein verification steps are as follows: Take 1mL of ind...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com