Probe set for detecting SARS-CoV-2 RNA, ECL biosensor and preparation method and application thereof
A biosensor, DNA probe technology, applied in biochemical equipment and methods, microbial determination/inspection, chemiluminescence/bioluminescence, etc., can solve problems such as increasing electron transfer distance, reagent consumption, affecting ECL intensity, etc. High electron transfer efficiency, good specificity and reproducibility, the effect of enhancing RuECL intensity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
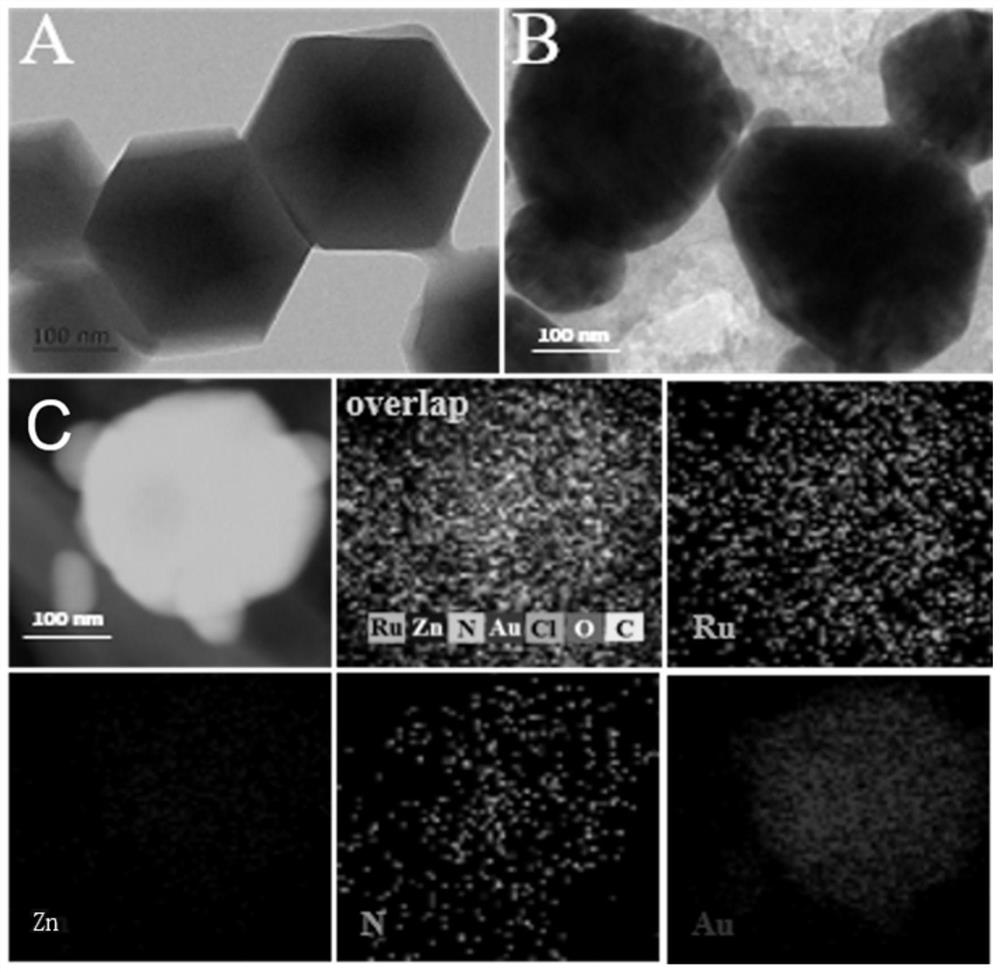
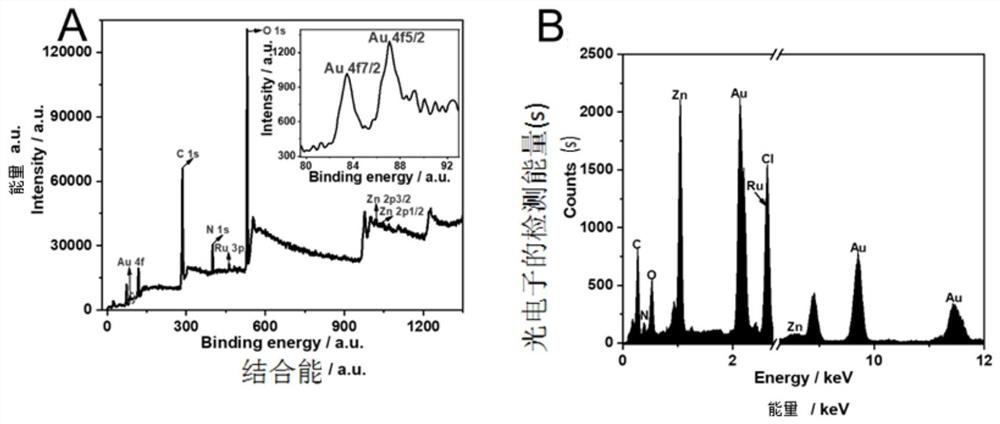
[0082] 1.1 Synthesis of Ru-PEI / Au@ZIF-8
[0083] 1.1.1 Synthesis of ZIF-8
[0084] 1) Zinc acetate dehydrating agent (Zn(AC) 2 -2H 2 O) heated to 55°C and kept for 45 minutes to remove moisture from the crystals to give Zn(AC) 2 powder;
[0085] 2) Add Zn(AC) 2 The powder (200mg) and 2-methylimidazole (295mg) were dissolved in methanol (22ml). The mixture was then diluted with water under continuous stirring to obtain a final concentration of 1 mmol Zn(AC) 2 and 3.3 mmol of 2-methylimidazole. When the mixture turns milky, age with stirring at room temperature for a day.
[0086] 3) The mixture was centrifuged, washed repeatedly with methanol and water, and then dried in an incubator to obtain a white powder, namely ZIF-8, which was used for the next step of modification.
[0087] Carry out amino modification to ZIF-8 surface, comprise the following steps:
[0088] 1) Sonicate 1.5 mg of ZIF-8 into 20 ml of N,N-dimethylformamide (DMF) to obtain a uniformly dispersed sus...
Embodiment 2
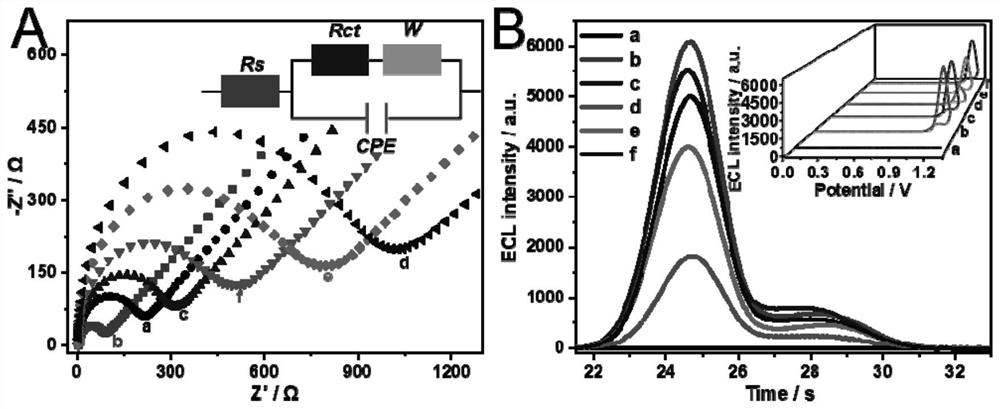
[0103] 2.1 The preparation of ECL biosensor, including the following steps:
[0104] S1: Before building the sensor, the GCE needs to be cleaned. A GCE with a diameter of 3 mm was immersed in a piranha solution (98% (v / v) H 2 SO 4 and 30% (v / v) H 2 o 2 The molar ratio is 4 to 1) to remove non-specific adsorbed substances on the electrode surface, and then repeatedly polished with 0.05 μm alumina powder and ultrasonically cleaned in ethanol and water until a bright and clean interface appeared. Then activate the electrode in sulfuric acid with a concentration of 0.1M, the voltage is -0.8-0.8V, and the scan rate is 0.1V / s until a continuous and stable CV peak appears. After rinsing with ultrapure water, the electrode was dried in N 2 Dry under the hood for the next step of biosensor construction.
[0105] S2: Drop 0.5% (mass percentage) of Nafion (N169478-5ml, Aladdin) and 8 μL of Ru-PEI / Au@ZIF-8 on the spot-free surface of GCE to obtain Nafion / GCE and Ru-PEI / Au in turn @...
Embodiment 3
[0125] Example 3 Detection of SARS-CoV-2 RNA using CRISPR-Cas 12a trans-cleavage properties and ECL biosensor
[0126] In order to prove the use of CRISPR-Cas 12a trans-cleavage characteristics in Example 2 and the detection level of SARS-CoV-2 RNA by ECL biosensors, this example analyzed the relationship between ECL intensity and target RNA concentration, and obtained their The mathematical relationship between them enables the possibility of inferring the target concentration from the signal intensity. Such as Figure 4 As shown in (A), when the concentration of SARS-CoV-2 RNA (SEQ ID: NO.5) in the sample increases, the ECL intensity gradually becomes stronger, which further confirms that the target RNA can drive the DSN enzyme to cycle with the CHA reaction process, Activate the activity of the CRISPR-Cas12a system to achieve the hydrolysis of Fc-labeled DNA probes. This embodiment also explored the linear relationship between ECL intensity and target RNA concentration, a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com