Antibacterial peptide prediction method and device based on protein pre-training representation learning
A prediction method and pre-training technology, applied in the field of computer identification of antimicrobial peptide components, can solve the problem that the model cannot be used universally
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
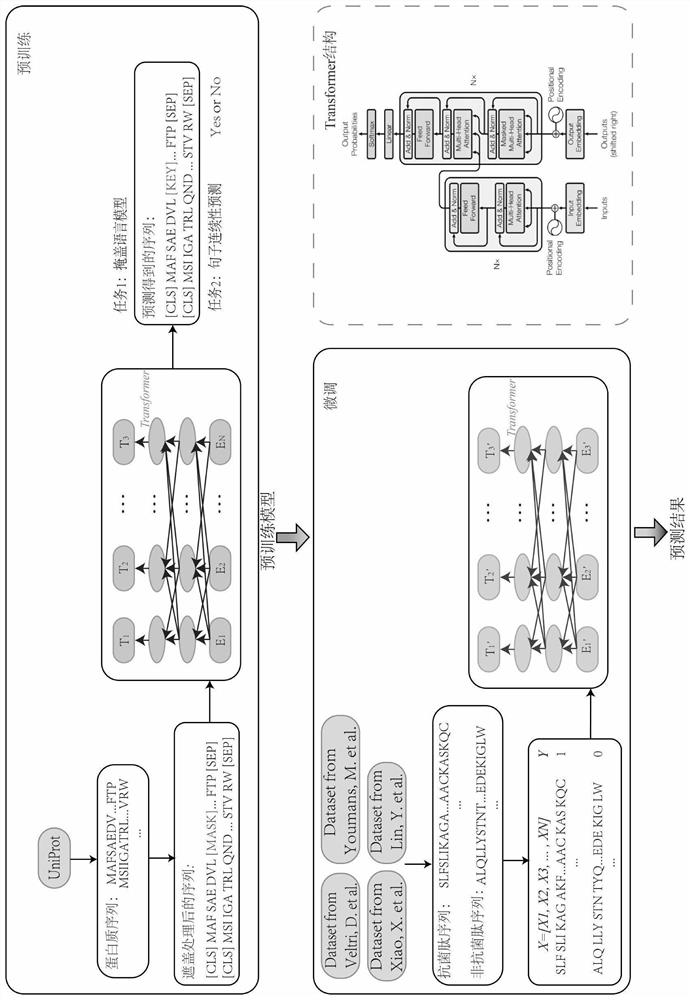
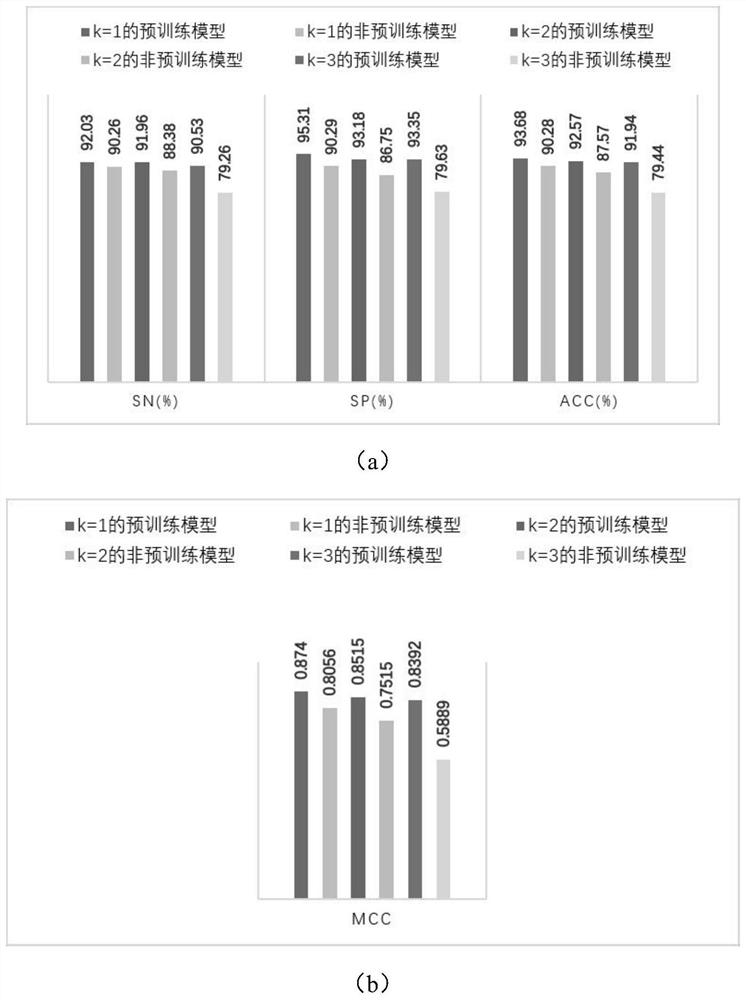
[0033] The antimicrobial peptide prediction method based on protein pre-training characterization learning of the present invention applies the method of pre-training + fine-tuning in the deep learning of natural language processing, including the following steps: first train a language model on a large amount of unmarked corpus, and then Fine-tuning the model using a specific data set with a small amount of data, such as a text classification data set, a named entity recognition data set, etc., usually takes only a short time. Therefore, after pre-training a model, the model can be quickly migrated to any natural language processing task, which helps the model save training time and computing resources. The number of experimentally determined antimicrobial peptides is far less than the number of proteins known to exist and sequenced. Using the antimicrobial peptide sequence to fine-tune the pre-trained model obtained from a large amount of protein data will help to mine riche...
Embodiment 2
[0104] Based on the same inventive concept as the antimicrobial peptide prediction method based on protein pre-training characterization learning in the foregoing embodiment 1, the present invention also provides a computing device, including one or more processors and memories, on which the There is a computer program, and when the program is executed by a processor, it realizes the steps of any one of the aforementioned methods for predicting antimicrobial peptides based on protein pre-training representation learning.
[0105] The computing device in this embodiment may be a general computer, a dedicated computer, a server or cloud computing, all of which are well known in the art.
[0106] Those skilled in the art should understand that the embodiments of the present invention may be provided as methods, systems, or computer program products. Accordingly, the present invention can take the form of an entirely hardware embodiment, an entirely software embodiment, or an embo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com