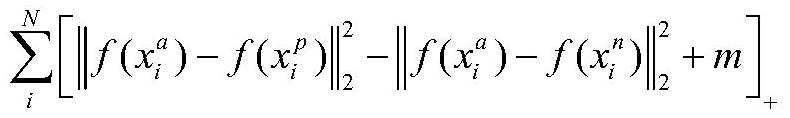Protein folding identification method based on triple loss
A technology of protein folding and identification method, which is applied to the analysis of two-dimensional or three-dimensional molecular structures, instruments, biological neural network models, etc., can solve the problems of "dimension disaster, indirect inefficiency, large dimension, etc., and achieve identification The effect of fast speed, faster recognition speed and higher accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
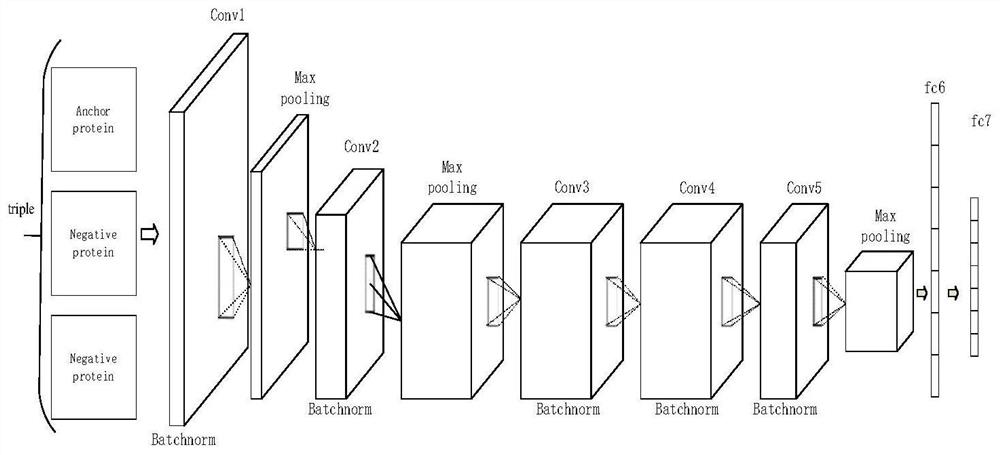
[0028] Such as figure 2As shown, a protein folding recognition method based on the loss of triplets first uses one-hot coding to encode the protein to obtain the digital expression of the protein sequence, and then inputs it into the SSA program to obtain the The contact graph of , for better expression, the contact graph between protein residues and residues is named RRcontact, and then RRcontact is used as input data, input into the pre-trained deep learning framework, and the output of the network is It is a protein-specific folding recognition feature, named f; finally, compare the query protein f(query) with the template protein f(template) of the known protein folding category in the protein database, and the nearest template protein to the query protein Fold classes are assigned to query proteins.
[0029] The process is described in more detail below in conjunction with the accompanying drawings:
[0030] Step 1: Training data preprocessing: Use one-hot encoding to ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com