Lactobacillus plantarum SD26 as well as product and application thereof
A technology of Lactobacillus plantarum and SD26, applied in the field of microorganisms, can solve the problems of aggravating bacterial resistance and destroying the balance of oral ecosystem, and achieve the effect of good application potential
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] The separation method of embodiment 1 lactic acid bacteria
[0060] Weigh 5g of kimchi sample and put it into a sterile homogeneous bag, add 45mL of sterile normal saline, beat and mix well, then dilute in gradient, and the dilution factor is 10 -3 、10 -4 、10 -5 、10 -6 100 μL of the sample, coated in 2.5% CaCO 3 MRS and M 17 On the plate, culture at 37°C for 24h. Colonies with good growth and obvious calcium-dissolving circles were picked, separated and purified repeatedly by plate streak separation method, and 9 strains of lactic acid bacteria were screened and isolated from kimchi samples, numbered SD23-SD31.
Embodiment 2
[0061] Embodiment 2 inhibits the screening of oral pathogenic bacteria lactic acid bacteria
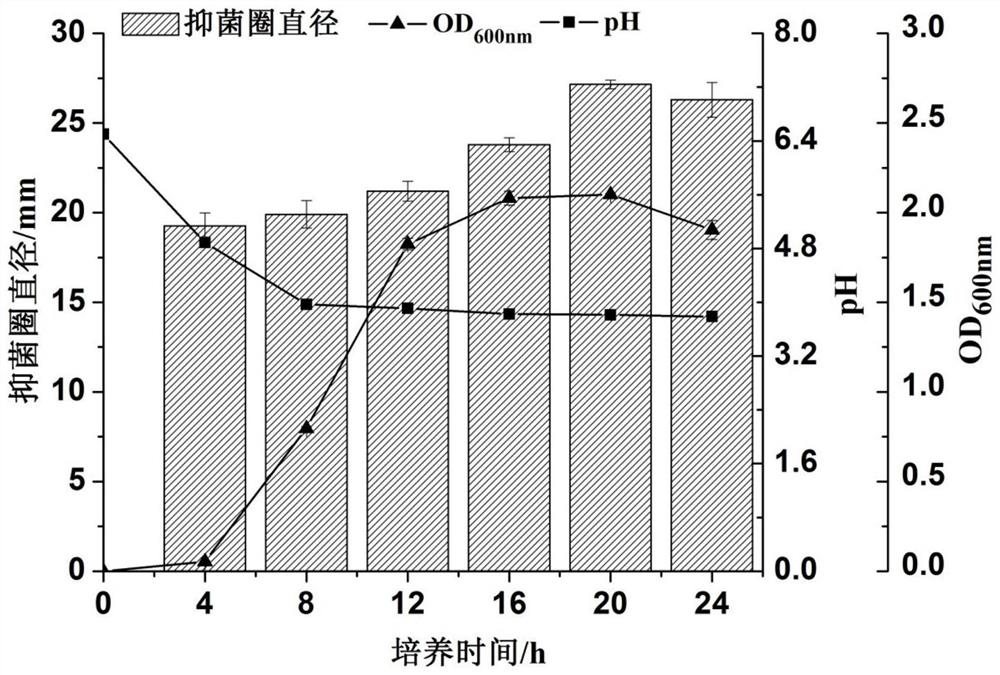
[0062] After the isolated lactic acid bacteria were inoculated on the MRS liquid medium for activation, they were inoculated in 5 mL of the MRS liquid medium with an inoculation amount of 2%, and cultured at 37° C. for 24 hours. After centrifugation at 10,000 r / min for 10 min, filter with a 0.22 μm sterile filter to obtain the cell-free fermentation supernatant. The strains with obvious inhibition zone against Streptococcus mutans, Candida albicans, Fusobacterium nucleatum and Porphyromonas gingivalis were screened by Oxford cup method. And MRS liquid medium was used as negative control (CK), and Nisin, potassium sorbate and natamycin were used as positive controls. Each test was repeated three times, and the diameter of the inhibition zone was measured with a vernier caliper using the cross method. The test results are shown in Table 2.
[0063] Table 2 The inhibitory activity of d...
Embodiment 316
[0067] Example 3 Identification of 16S rRNA
[0068] Genomic DNA of unknown strains was extracted according to the instructions of the bacterial genomic DNA extraction kit, and PCR amplification of the 16S rRNA gene was performed using the genomic DNA as a template. As amplification primers, universal primers 27F (5'-AGAGTTTGATCCTGGCTCAG-3') and 1492R (5'-TACGGCTACCTTGTTACGACTT-3') were used. The PCR reaction system is shown in Table 3.
[0069] Table 3 PCR reaction system
[0070]
[0071] PCR reaction conditions: pre-denaturation at 95°C for 5min; denaturation at 95°C for 30s, annealing at 55°C for 30s, extension at 72°C for 30s, 34 cycles; final extension at 72°C for 5min.
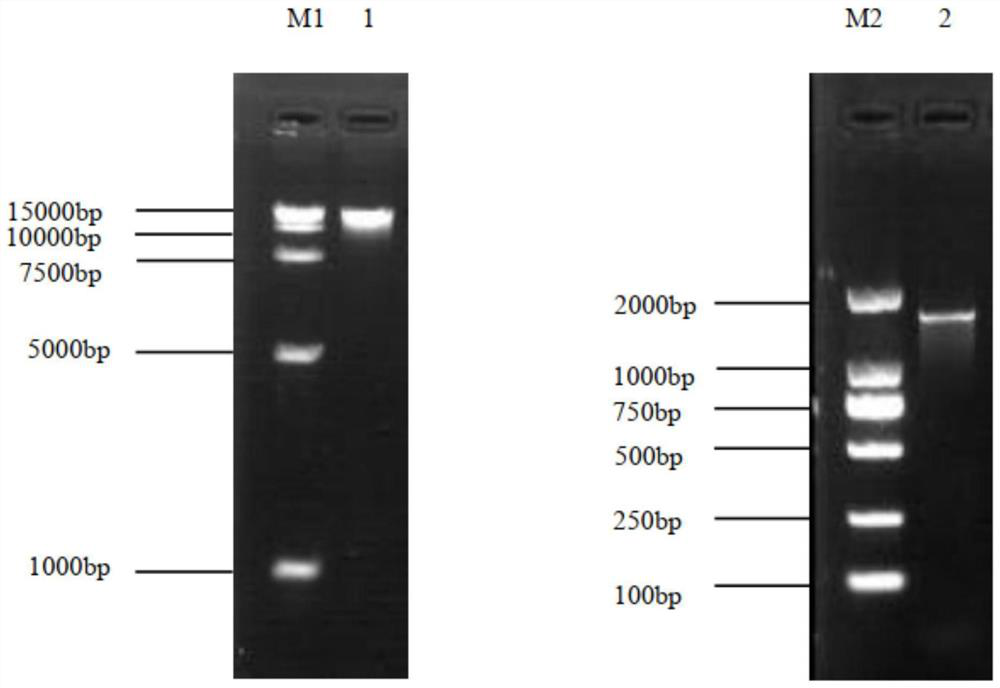
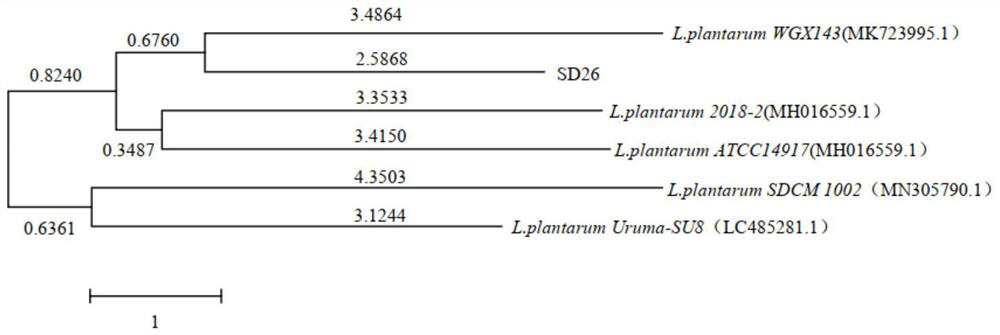
[0072] After the extracted genome and PCR amplification products were analyzed and detected by agarose gel electrophoresis, the PCR products were subjected to DNA sequencing (Shanghai Sangon Bioengineering Co., Ltd.). The sequencing sequence results were searched for approximate sequences in the N...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com