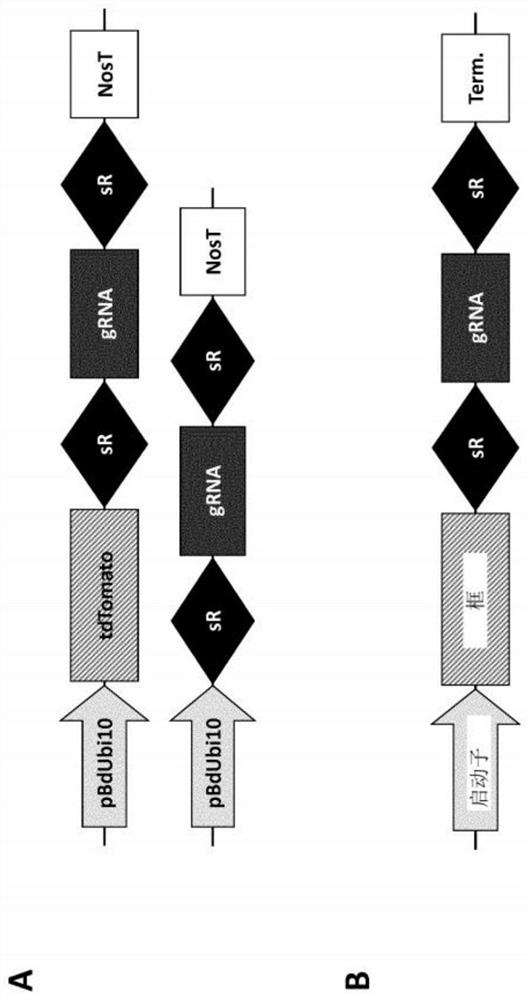
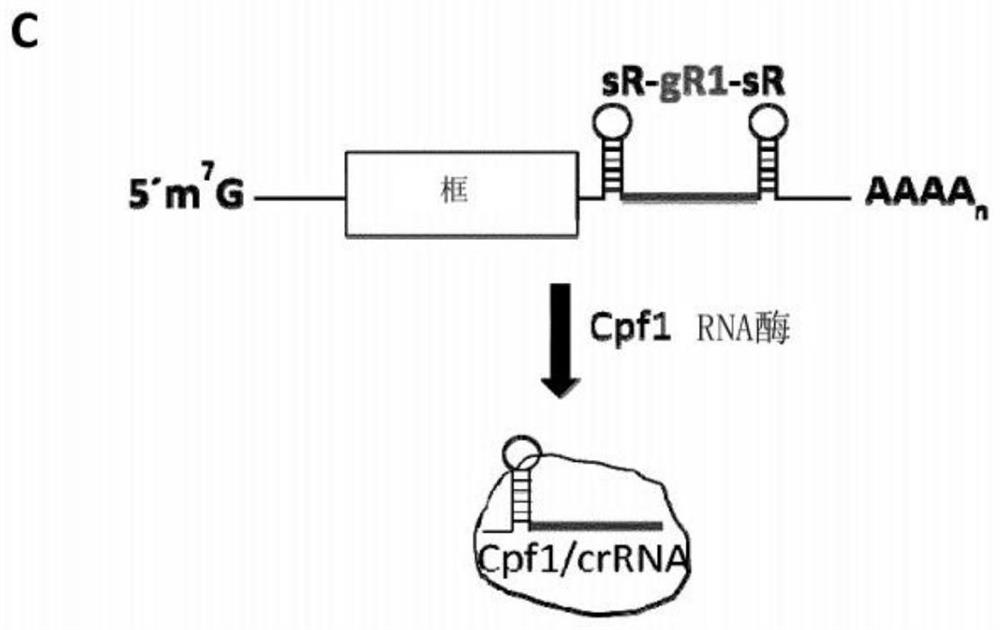
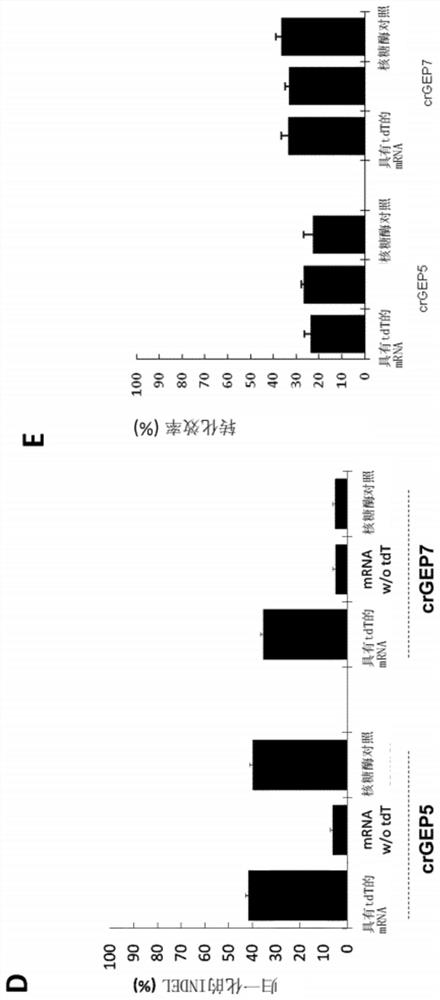
Optimized plant CRISPR/CPF1 systems
A plant and delivery system technology, applied in the field of optimized systems and plant delivery systems, can solve the problem of low predictability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0259] Example 1: Target Sequence
[0260] For the purpose of this analysis, various gene editing plasmids (GEPs) were designed and used. As shown below, Table 1 provides the internal designation (crGEP) of the different gene editing plasmids (left column), information about the target gene (middle column), and the respective target sites of selected crRNAs or sgRNAs (right column). The target site represents the actual binding site in the genomic target region or gene of interest.
[0261] Table 1:
[0262]
[0263]
Embodiment 2
[0264] Embodiment 2: plant protoplast transfection
[0265] When transfecting plant protoplasts for the experimental purposes disclosed herein, protocols known in the art were used according to the following steps:
[0266]The buffers and solutions used are enzyme liquid, enzyme wash, enzyme wash buffer (EWB), MMG (ethylene glycol-magnesium mannitol) buffer, etc., which contain 0.1 to 0.5M mannitol, 15mM to 20mM magnesium chloride and 4 mM MES (pH 10 to 40% PEG (polyethylene glycol) calcium, stop buffer and W5 buffer (for example, containing 154 mM sodium chloride, 125 mM calcium nitride, 5 mM potassium chloride, 2 mM M ES (pH 5.7)) glucose ).
[0267] First, 20 μg (or unless otherwise specified) of plasmid DNA was added to a 2 ml tube at 4°C. Second, harvest fully expanded first and second leaves from 10-14-day-old etiolated seedlings in a true-leaf greenhouse and place them in a bag with wet paper towels. The leaves were cut into thin strips weighing 4.5 g. Place in a de...
Embodiment 3
[0272] Example 3: Next Generation Sequencing (NGS) Protocol
[0273] When using next-generation tests for experiments, the following protocols are followed: Library preparation: Libraries are prepared by two PCR steps to amplify regions of interest and add sequencing adapters. Barcodes were designed with primers and added in the first PCR step for sample identification. Adapters are added during the second PCR for sequencing. Next-generation sequencing (NGS): The amplicons were sequenced using the Illumina Miseq150 PE platform. The sequencing coverage was 100,000-fold for the protoplast population, and 250,000-fold, 300,000-fold, and 50,000-fold for immature embryos and callus, Agrobacterium-transformed leaf samples, and bombarded wheat leaf samples, respectively. Analyze data using FastQC+Jemultiplexer+Trimmomatic for read QC and demultiplexing, use CRISPResso for identification of target gains and losses, and edit event calls via internal impact client scripts. The entire...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com