Method and kit for detecting sperm DNA fragment rate
A fragmentation rate and kit technology, applied in the field of in vitro diagnostic reagents, can solve problems such as complex operation process, incomplete enzymatic hydrolysis, complex methods, etc., and achieve accurate and repeatable results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
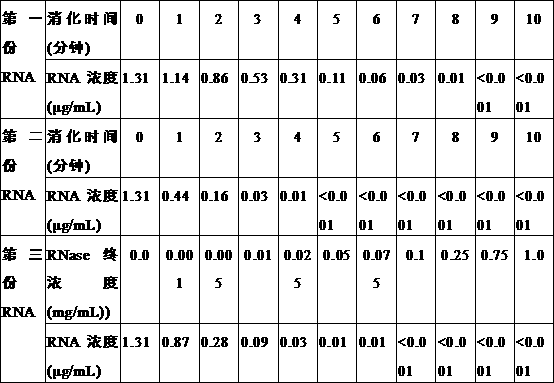
[0056] The present embodiment provides the use of the method of the present invention to detect sperm DNA fragmentation rate, wherein the detection of RNase enzyme digestion effect comprises the following steps:
[0057] 1.1 Preparation of items:
[0058] Prepare sterile, disposable plastic products (centrifuge tube, tip), Trizol reagent, chloroform, isopropanol, 75% ethanol, RNase-free water, RNase A, nucleic acid quantitative detector.
[0059] 1.2 Extraction and isolation of total RNA from sperm cells:
[0060] 1.2.1 Lysis of sperm cells (cell volume 5×10 8 indivual )
[0061] The liquefied semen was centrifuged at 800g for 10 minutes at room temperature, and the supernatant was discarded completely. Add 1mL Trizol and immediately mix by pipetting 5 times.
[0062] 1.2.2 Let stand at room temperature for 5 minutes. Then add 0.25mL of chloroform, and vigorously invert the centrifuge tube to mix. After standing for 5 minutes, centrifuge at a centrifugal speed of 12000g ...
Embodiment 2
[0077] This example provides the detection of sperm DNA fragmentation rate using different methods.
[0078] Specifically, the following steps are included:
[0079] 2. Sample Preparation
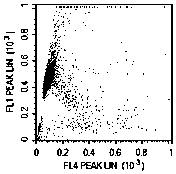
[0080] 2.1 Divide 20 naturally liquefied semen (each sample volume is 1.5-5.5mL, sperm concentration is 15-200 million / mL) into 3 parts, and use the following 3 methods to detect sperm DFI: method A: flow cytometry of digested RNA Cytometry to detect DFI; method B: flow cytometry to detect DFI after undigested RNA; method C: sperm chromatin diffusion method to detect DFI;
[0081] 2.2 Method A detects DFI:
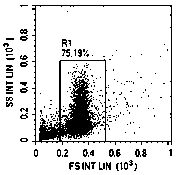
[0082] 2.2.1 Dilute sperm with 37°C buffer to 1-2×10 6 pcs / mL; the buffer solution is a mixture of 0.15 mol / L NaCl (sodium chloride), 1 mmol / L EDTA (ethylenediaminetetraacetic acid), pH 7.4;
[0083] 2.2.2 Add 500 μL of diluted sperm into the sample tube of the flow cytometer, add 5 μL of 10 mg / mL RNase A (purchased from Sigma), and digest at 37°C for 5 minutes;
[0084] After 2.2....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com