A kind of mnep monomer variant and its application
A variant and monomer technology, applied in the field of characterization of target polynucleotides, can solve the problems of few types of porins, the application of Mnep monomer variant is not mentioned, etc., and achieve the effect of high sequencing accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0198] Embodiment 1 Preparation of Mnep monomer variant
[0199] 1. Plasmid construction
[0200] The protein sequence of the Mnep monomer variant is optimized by codons corresponding to amino acids, and appropriate restriction endonuclease sites are added at both ends of the gene. Specifically, the NcoI site ccatgg is added to the 5' end, and the xhoI site is added to the 3' end. Click ctcgag, and then perform gene synthesis, and the synthesized gene is cloned into the expression vector pET24b.
[0201] 2. Site-directed mutagenesis of the target gene to prepare the nucleotide sequence of the Mnep monomer variant
[0202] Induced mutation gene (PCR reaction) Use the plasmid to be mutated as a template, and use the designed primers and KOD plus high-fidelity enzyme to perform PCR amplification reaction to induce the mutation of the target gene.
[0203] Specific steps are as follows:
[0204] 1. Design point mutation primers, prepare template plasmid DNA (plasmid DNA contain...
Embodiment 2
[0231] The preparation of embodiment 2 porins
[0232] 1. Take 12 μL of Mnep-K0 BL21 (DE3) glycerol and inoculate it into 12 mL (1:1000) of fresh LB medium containing a final concentration of 50 mg / mL kanamycin, and activate overnight at 37 ° C by shaking at 200 rpm; the next day press 1 % inoculum size Expand the culture to 2 L of LB medium containing a final concentration of 50 mg / mL kanamycin. 37℃, 220rpm culture to OD 600 After = 0.6-0.8, the ice bath was cooled rapidly, and then IPTG was added to the culture system to a final concentration of 1 mM, and the expression was induced overnight at 18° C. and 220 rpm.
[0233] 2. The next day, collect the cells by centrifugation at 6000rpm at 4°C for 15 minutes, resuspend the cells according to the ratio of cells: lysis buffer = 1:10 (m / v), then add mixed protease inhibitors, and crush under high pressure until the cells become clear .
[0234] 3. Add 1% OPOE and 0.1% FC12, stir and solubilize at room temperature for 1-2 hours....
Embodiment 3
[0238] The sequencing application of embodiment 3 porins
[0239] In buffer (400mM KCl, 10mM HEPES pH 8.0, 50mM MgCl 2 ), a single nanopore was inserted into a phospholipid bilayer and electrical measurements were obtained from a single nanopore.
[0240] Specific steps are as follows:
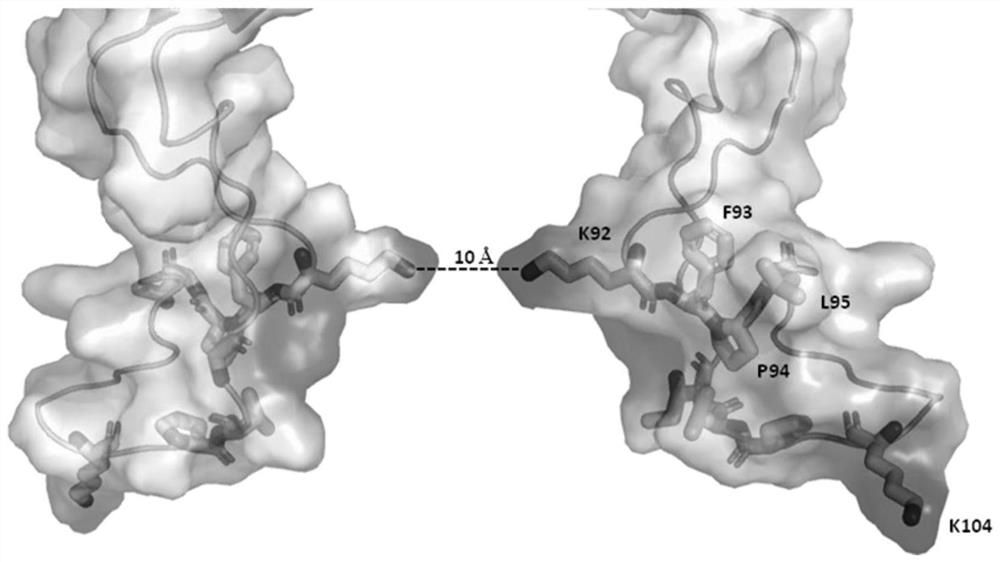
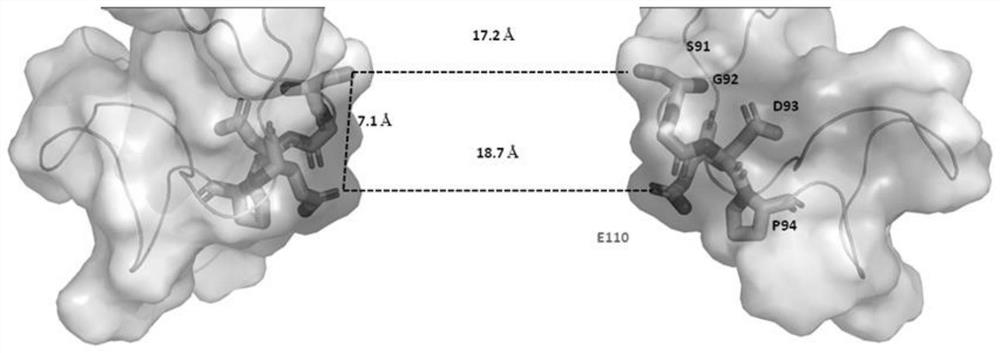
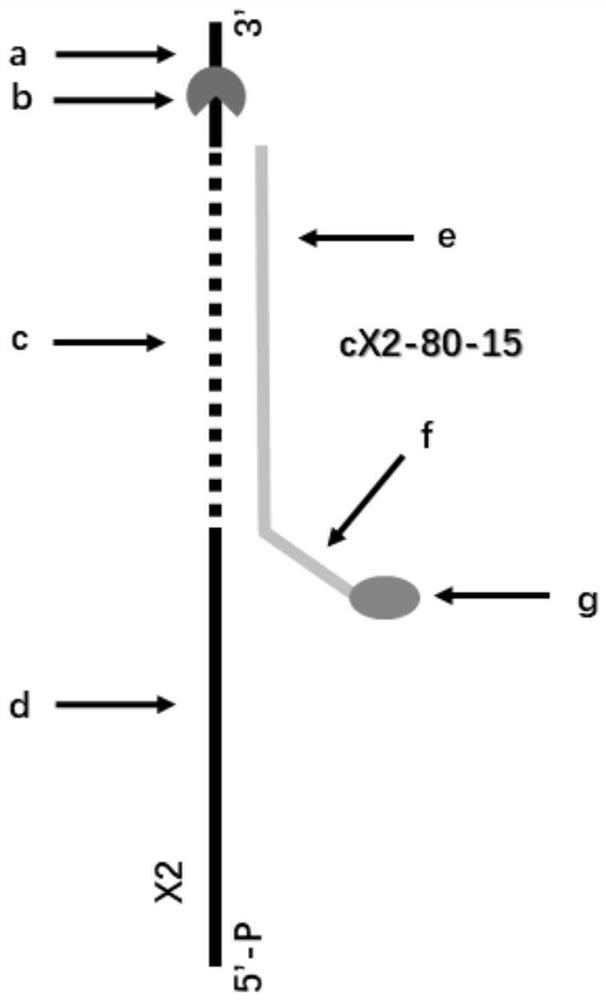
[0241] In the single porin (Mnep-K0 nanopore of SEQ ID NO: 1 whose amino acid sequence is mutated to G92K / D93F / G95L / A104K, the stick model is as figure 1 shown) after inserting into the phospholipid bilayer, the buffer (400mM KCl, 10mM HEPES pH 8.0, 50mM MgCl 2 ) through the system to remove any excess Mnep-KO nanopores. Add the DNA construct X2&cX2-80-15 or S1T&S1MC (1-2nM final concentration) into the Mnep-K0 nanopore experimental system, after mixing, make the buffer solution (400mM KCl, 10mM HEPES pH 8.0, 50mM MgCl 2 ) through the system to remove any excess DNA constructs X2&cX2-80-15 or S1T&S1MC. Then helicase (EF8813-1, 15nM final concentration), fuel (ATP3mM final concentration) p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com