Novel mutant protein for improving malic acid yield
A mutant protein, malic acid technology, applied in the biological field, can solve problems such as cell burden
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0194] Preparation of mutant proteins
[0195] The present invention also provides a method for preparing the pyruvate carboxylase mutant protein malate transporter mutant protein, comprising culturing the host cell of the present invention under suitable expression conditions, thereby expressing the pyruvate carboxylase mutant protein and / or the the malate transporter mutein; and
[0196] The pyruvate carboxylase mutein and / or the malate transporter mutein are isolated.
[0197] The mutein obtained may also optionally be purified to obtain a more pure mutein product.
[0198] Preferably, the conditions suitable for expression include conventional techniques in the art, and purification techniques include nickel column purification, ion exchange chromatography, and the like.
[0199] carbon source
[0200] The carbon source that can be used in the present invention is not particularly limited, it is a type of nutrient for the growth of microorganisms (such as the host cell ...
Embodiment 1
[0209] Example 1 Pichia pastoris dicarboxylic acid transporter screening platform construction
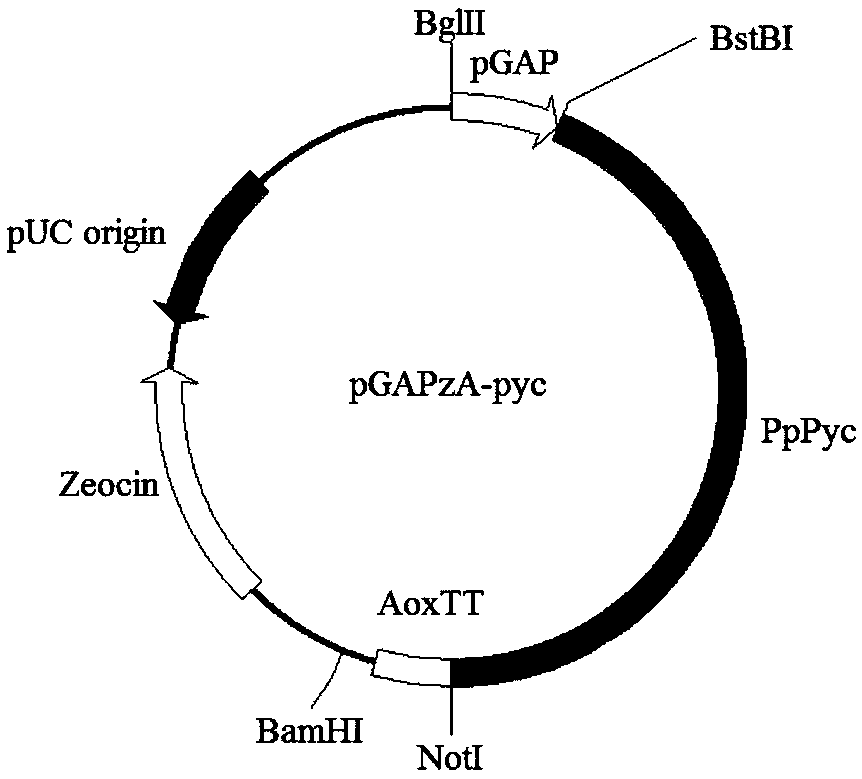
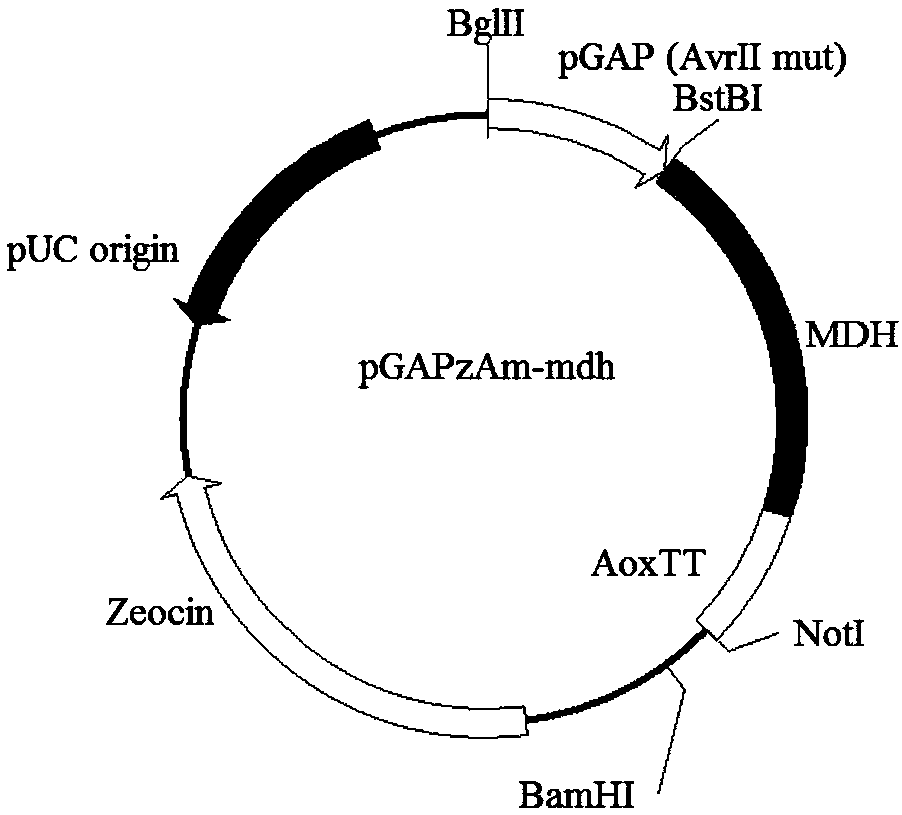
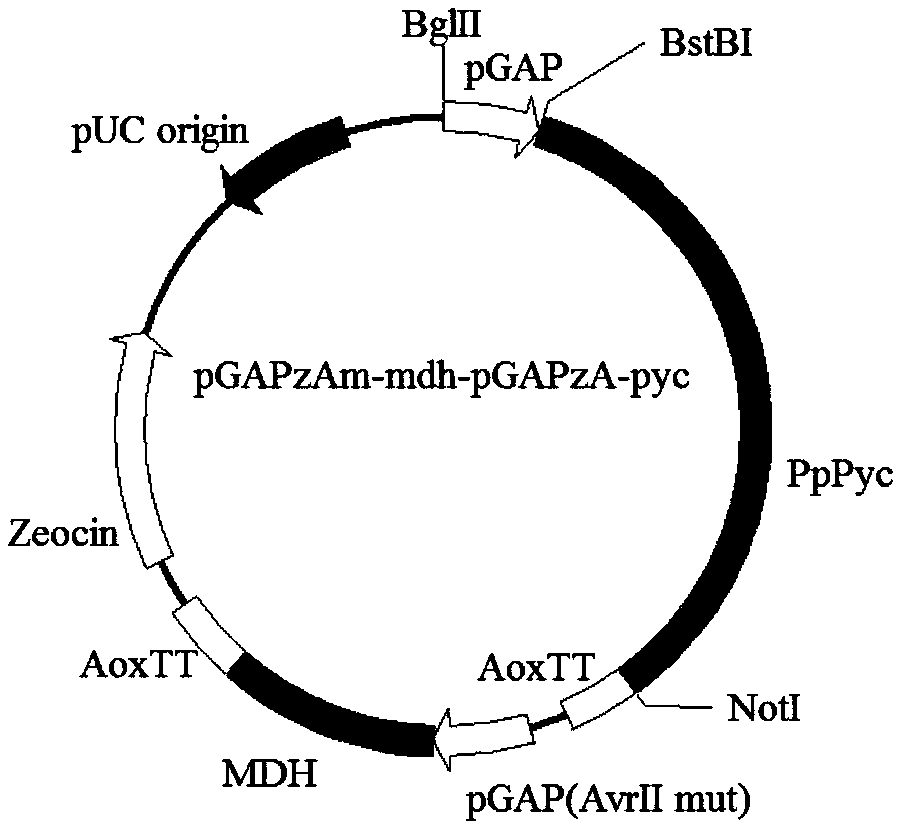
[0210] 1. Construction of vectors for overexpressing pyruvate carboxylase and malate dehydrogenase in Pichia pastoris:
[0211] Use primers AvrII-F (SEQ ID NO: 44): (CCACCGCCCGTTACCGTCCGAAGGAAATTTTACTCTGCTGGAG) and AvrII-R (SEQ ID NO: 45): (CTCCAGCAGAGTAAAATTTCCTTCGGACGGTAACGGGCGGTGG) to perform PCR with pGAPzA as a template. The PCR reaction system is: 5×phusion HF buffer 10 μl 10 mM dNTPs 1 μl, AvrII-F 1 μl, AvrII-R 1 μl, pGAPzA 1 μl, Phusion DNA polymerase 0.5 μl, water 35.5 μl. The PCR reaction conditions are as follows: first 98°C for 30s; then 98°C for 10s, 60°C for 30s, 72°C for 3min, 30 cycles; finally 72°C for 10min, 4°C for 10min. After the PCR reaction, the product was purified through a purification column, and 1 μg of plasmid was added to 1 μl of restriction endonuclease DpnI to digest and remove the template, and then heat inactivated at 85°C for 5 minutes, and 5 μl ...
Embodiment 2
[0222] Example 2 Construction and screening of dibasic carboxylic acid transporter mutation library.
[0223] 1. Construction of expression vector of dicarboxylic acid transporter:
[0224] Primers are:
[0225] Hph‐BamH1 SEQ ID NO:50GGGGGATCCTGTACAGCTTGCCTCGTCCCC
[0226] Hph‐R SEQ ID NO:51GTCGACACTGGATGGCGGCGTTAG
[0227] Using the pAG34 plasmid as a template, the hygromycin gene was amplified by PCR. The PCR reaction conditions were: first 98°C for 30s; then 98°C for 10s, 60°C for 30s, 72°C for 2min, 30 cycles; finally 72°C for 10min, 4°C for 10min. After the PCR reaction, the product was purified through a purification column, digested with BamH1, and connected to the pGAPzA plasmid digested with BamH1 and EcoRV, and the ligated product was transformed into E. coli competent cell DH5α. The monoclonal sequencing was correct, and pGAP-hph was obtained .
[0228] Primers are:
[0229] his‐BglII SEQ ID NO:52GGGAGATCTGTTGTAACACTGGCAGAGCATTACG
[0230] his‐BamH1 SEQ ID NO:...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com