Specific primer and method for detecting towneri acinetobacter and application
A specific, bacilli-based technology, applied in the field of microbiology, can solve problems such as nucleic acid and primers that have not been invented by anyone, and achieve the effects of simple detection, easy determination, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1 Establishment of a specific PCR detection method for Acinetobacter towneri
[0029] (1) Design of specific primers for Acinetobacter towneri
[0030] In this embodiment, for the outer membrane protein A gene (OmpA gene) of Acinetobacter towneri, a pair of specific primers with strong specificity and high sensitivity to Acinetobacter towneri are designed. The specific information of the specific primer pair is as follows :
[0031] The primer sequences are:
[0032] F: 5'-GACTGAGCCCGTAGAAGA-3';
[0033] R: 5'-ACAGTTGATTACCCACCC-3'.
[0034] (2) DNA template preparation of Acinetobacter towneri
[0035] 2.1 Extraction of Acinetobacter towneri DNA
[0036] Utilize TE buffer boiling method to extract the DNA of Acinetobacter towneri, concrete steps are as follows:
[0037] 1) Take 1 mL of Acinetobacter towneri cultured in a 37°C constant temperature incubator for 24 hours in a 4 mL centrifuge tube, centrifuge (room temperature, 6000r, 3 min), and discard the...
Embodiment 2
[0050] Embodiment 2 Specificity evaluation test
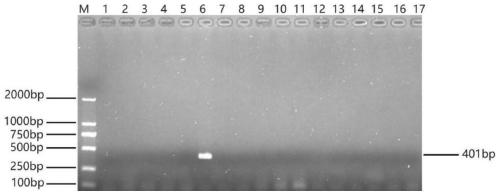
[0051] According to the DNA template preparation and PCR detection method in Example 1, the Acinetobacter baumannii marcescens, Acinetobacter Pitt, Acinetobacter Johnson, Morganella, Staphylococcus aureus, E. Reedia, Klebsiella pneumoniae, Neisseria, Pasteurella, Shigella, Pseudomonas aeruginosa, Proteus vulgaris, Citrobacter kirsi, Serratia marcescens for PCR amplification Increased response.
[0052] Depend on figure 2 In the electrophoresis results, it can be seen that only Acinetobacter towneri has a clear and bright specific band at 401bp, while other bacteria have no specific band.
Embodiment 3
[0053] Embodiment 3 sensitivity evaluation test
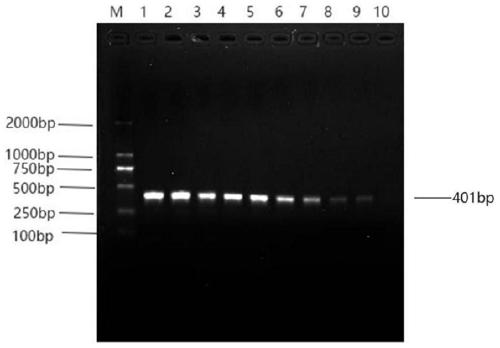
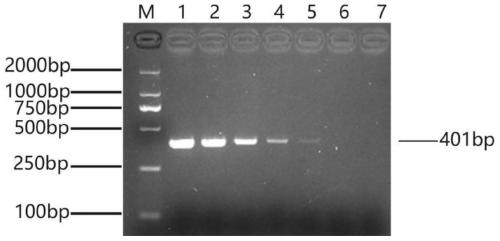
[0054] Inoculate Acinetobacter towneri in 1mL nutrient broth liquid medium, place it in a 37°C constant temperature incubator and culture for 24 hours to enrich the bacteria, then use 10-fold gradient dilution with sterilized nutrient broth liquid medium, and count the bacterial concentration by plate method 8.7×10 9 CFU / mL, get 1mL bacterium liquid and extract genomic DNA according to embodiment 1, press 10 with sterilized double distilled water 0 -10 -6 Perform 10-fold gradient dilution of genomic DNA, use 7 gradient dilutions as templates for PCR amplification, detect the amplified products by gel electrophoresis, and observe the results of gel electrophoresis under ultraviolet light.
[0055] Depend on image 3 It can be seen that a clear band can be seen in lane 4, and the corresponding detection bacteria concentration is 8.7×10 6 CFU / mL, this method has good sensitivity.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com