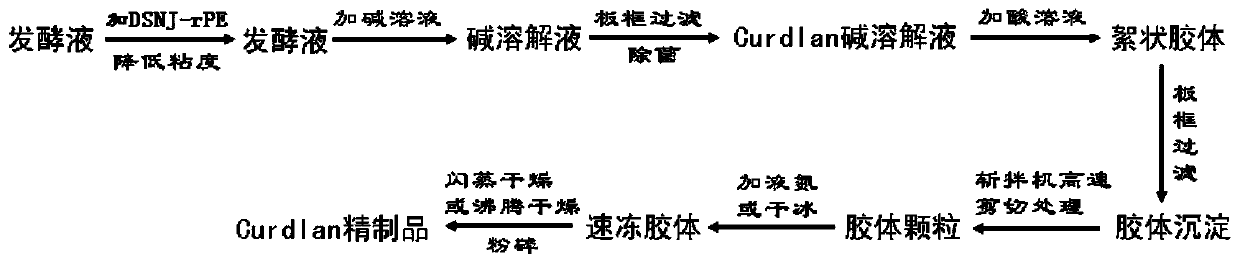
Efficient post-extraction method of curdlan
An extraction method and polysaccharide technology, which are applied in the field of high-efficiency post-extraction of available polysaccharides, can solve the problems of colloid loss, large usage of organic solvents, long drying time, etc., and achieve improved recovery efficiency, reduced production cost, and improved drying efficiency. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1: Cloning and sequencing analysis of protease DSNJ-rPE gene
[0039] Search several protease sequences from NCBI website, carry out bioinformatics analysis, use CODEHOP (Consensus-degenerate hybrid oligonucleotide primers) program to select two highly conserved amino acid sequences and design degenerate primers.
[0040] Using the soil genomic DNA extraction kit from MP bio ( Spin Kit for Soil) to extract the total genomes of 50 soil samples from different sources.
[0041] In the 50μl system, add upstream primer: 5′-gcaccggattccgacgacag-3′, and downstream primer: 5′-tcgccgcaggccgactctag-3′, the final concentration is 0.8μM, the final concentration of dNTPs is 0.2mM, and the total DNA of soil genome is 10ng, 2U Pfu DNA polymerase. The amplification program was 94°C for 5min; 35×(94°C for 30s, 55°C for 60s, 72°C for 60s); 72°C for 10min. The PCR products were sent to Shanghai Sangon Bioengineering Company for sequencing.
[0042] Analyzing the sequencing r...
Embodiment 2
[0043]Embodiment 2: the construction of prokaryotic expression vector of protease DSNJ-rPE gene (attached figure 1 )
[0044] According to the MTG gene sequence obtained by sequencing, SacI recognition sequence (GAGCTC) and XhoI recognition sequence (CTCGAG) were added to the 5' and 3' ends respectively, and Shanghai Sangon Bioengineering Company was commissioned to synthesize the gene. The synthesized gene and vector pET-23a were digested with appropriate amount of the same restriction endonuclease BamHI and ligated with T4 DNA ligase. The recombinant vector transformed Escherichia coli DH5α. Randomly pick a single colony from the transformation plate, insert it into LB liquid medium, shake culture, extract a small amount of plasmid, electrophoresis, use the plasmid with electrophoresis delay as a template for PCR verification, and send it to Shanghai Sangon Bioengineering Co., Ltd. after confirming that the connection is successful sequencing.
Embodiment 3
[0045] Embodiment 3: the expression of protease DSNJ-rPE gene in Escherichia coli
[0046] Transform the recombinant expression plasmid pET-23a-DSNJ-rPE containing DSNJ-rPE gene into Escherichia coli expression host strain BL21(DE3)pLysS, culture at 37°C for 10-11 hours, pick small colonies, and insert them into 50ml LB containing ampicillin Liquid medium, culture at 30°C at 70-90rpm overnight, take the seed solution according to the volume ratio of 1:40 and add it to 100ml LB liquid medium containing ampicillin, shake at 180rpm at 35°C for 2-3 hours until OD600 is about 0.6, add IPTG (final concentration 100 μg / ml) induction. After 1.5 hours, the cells were collected by centrifugation. Break up the bacteria, centrifuge to collect the supernatant, and obtain the recombinant DSNJ-rPE protease crude enzyme solution.
[0047] Recombinant protease DSNJ-rPE activity assay method: Dissolve fluorescently labeled casein in 0.2mL phosphate buffer (pH 7.0), mix well, absorb 50μL of su...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com