Polysaccharide cleavage monooxygenase encoding gene and enzyme thereof, and preparation method and applications
A technology of monooxygenase and polysaccharide, which is applied in polysaccharide cleavage monooxygenase coding gene and enzyme preparation and application field, can solve the problems such as no literature or patent report of gene and amino acid sequence, and achieve efficient degradation effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Example 1 Construction of polysaccharide cleavage monooxygenase MtLPMO9E eukaryotic expression strain
[0062] After performing multiple sequence alignment analysis on the polysaccharide cleavage monooxygenase gene sequence in The National Center for Biotechnology Information (NCBI) database, the polysaccharide cleavage monooxygenase of Myceliophthora thermophila was prepared to be synthesized. The coding region of the target gene MtLPMO9E in the Myceliophthora thermophila genome is 741bp long, its nucleotide sequence is shown in SEQ ID NO1, encoding 246 amino acids and a stop codon, its amino acid sequence is shown in SEQ ID NO 2, and its theoretical molecular weight It is 26.04KDa. The MtLPMO9E signal peptide is predicted to be 1-17 amino acids, the molecular weight is predicted to be 24.29KDa after removing the signal peptide, and the predicted isoelectric point is 6.03. The amino acid encoded by MtLPMO9E contains an AA9 (GH61) family domain. Through codon optimiza...
Embodiment 2
[0064] Example 2 Heterologous expression and purification of polysaccharide cleavage monooxygenase MtLPMO9E in Pichia pastoris
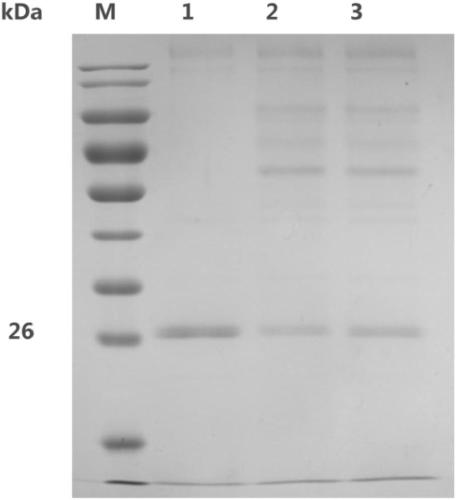
[0065] The polysaccharide cleavage monooxygenase MtLPMO9E was induced and expressed according to the method of Pichia pastoris expression manual. Polyacrylamide gel electrophoresis was used to detect the expression of polysaccharide cleavage monooxygenase LPMO9E, and the results were as follows: figure 2 shown. The BIO-PAD protein purification system was used to purify the target protein according to the isoelectric point of the protein. The purified polysaccharide cleavage monooxygenase LPMO9E showed a single band on the electrophoresis gel, and its position was consistent with the predicted molecular weight.
Embodiment 3
[0066] Example 3 Analysis of substrate binding activity of polysaccharide cleavage monooxygenase MtLPMO9E
[0067] Use affinity incubation and SDS-PAGE techniques to study the binding ability of MtLPMO9E to insoluble substrates: MtLPMO9E was incubated with the pretreated and washed substrate for 24 hours under buffer conditions, centrifuged to obtain the supernatant and precipitate, and the precipitate needed to be washed twice. SDS-PAGE was used to detect the proportion of MtLPMO9E protein in the upper layer and the precipitate to determine its binding ability, and it was found that: MtLPMO9E can bind to cellulose substrates (see image 3 ), MtLPMO9E has no binding activity with chitin substrates (see Figure 4 ).
[0068] Using isothermal calorimetry (ITC) to study the binding ability of MtLPMO9E to soluble substrates: MtLPMO9E and xyloglucan were equilibrated overnight in the same buffer, and titrated according to the operating procedure of the isothermal calorimeter. The ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com