Construction method and application of recombinant bacterium for producing chitosanase
A technology for producing chitosanase and chitosanase, applied in the biological field, can solve the problems of increased energy consumption and cost, and achieve the effects of high-efficiency expression, excellent hydrolysis characteristics, and good application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
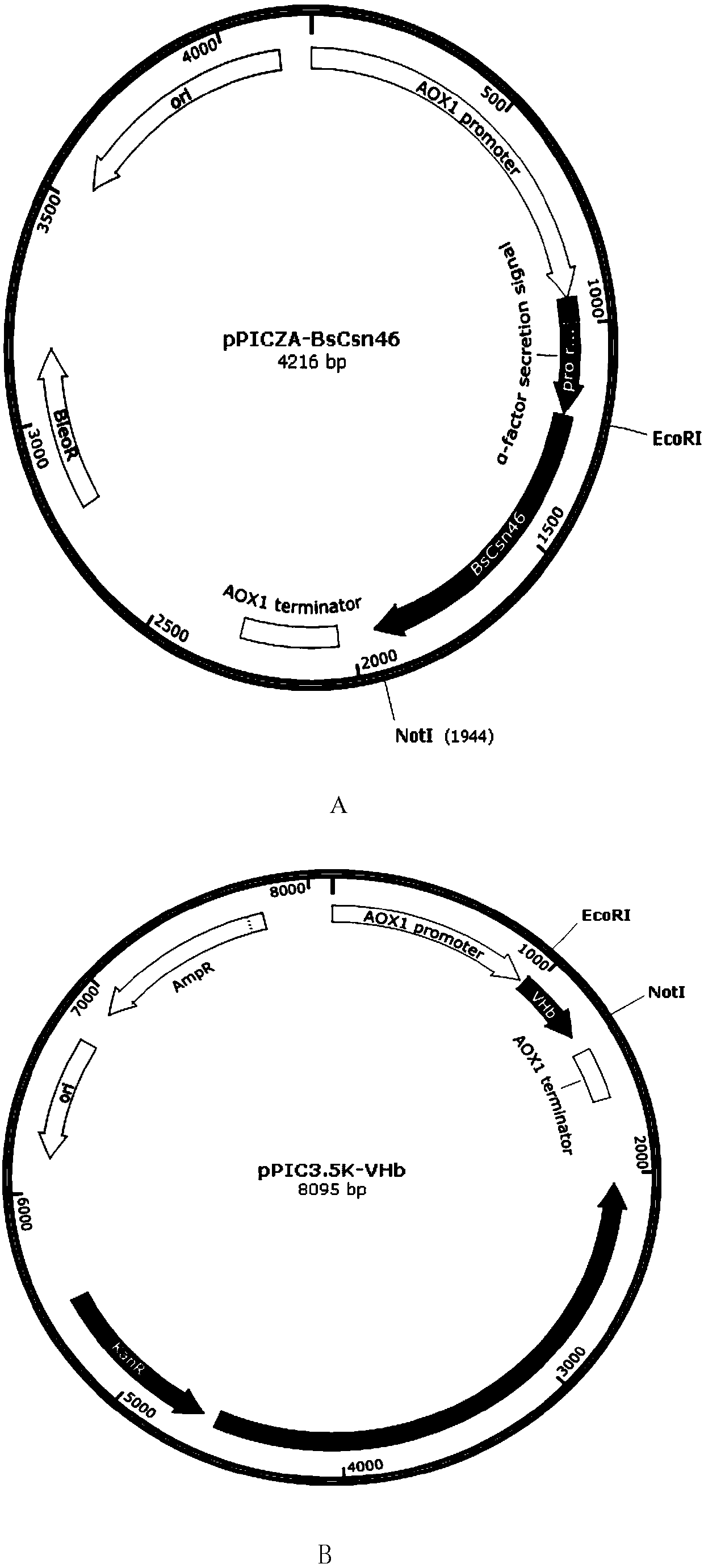
[0066] Embodiment 1, the preparation of recombinant bacteria GS115 / BsCsn46 / VHb
[0067] 1. Using the Bacillus subtilis WY-34 genome as a template, PCR amplification was performed using primers BsCsn46-F and BsCsn46-R to obtain the Bacillus subtilis chitosanase gene BsCsn46. The primer sequences are as follows:
[0068] BsCsn46-F: AACCG GAATTC ATGGGACTGAATAAAGATCAAAAGC (the underline is the EcoRI restriction site);
[0069] BsCsn46-R:AAGAAT GCGGCCGC TTATTTGATTACAAAATTACCGTAC (the underline is the NotⅠ restriction site).
[0070] The Bacillus subtilis chitosanase gene BsCsn46 was codon-optimized, and the optimized BsCsn46 gene sequence was shown in Sequence 1. Using the codon-optimized sequence as a template, primers BsCsn46-F and BsCsn46-R were used for PCR amplification. increase to obtain PCR products.
[0071] 2. The PCR product obtained in step 1 and pPICZA were digested with EcoRI and NotI respectively, and ligated with T4 DNA ligase at 16°C to construct the recombi...
Embodiment 2
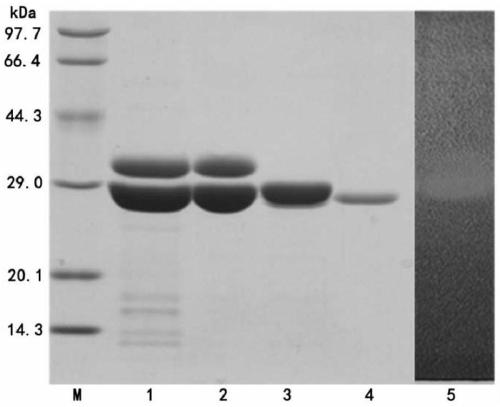
[0081] Embodiment 2, preparation and purification of recombinant chitosanase (BsCsn46)
[0082] 1. Preparation of recombinant chitosanase (BsCsn46) by high-density fermentation of recombinant bacteria
[0083] The high-density fermentation of the recombinant bacteria and the medium refer to the method in the Pichia Fermentation Process Guideline (Invitrogen), and the specific steps are as follows:
[0084] 1. Inoculate the recombinant bacterium that produces chitosanase activity higher (415U / mL) obtained by screening in Example 1 to 150mLYPD medium (tryptone 20g / L, yeast extract 10g / L, glucose 20g / L) , 30°C, 200rpm to OD 600 About 10, as a seed solution.
[0085] 2. Select a 5L fermenter (with a working volume of 1.5L) as a high-density fermentation vessel, add 150 mL of the seed liquid obtained in step 1 into a fermenter containing 1350 mL of fermentation medium for batch fermentation. During the fermentation process, 28% ammonia water was used to adjust the pH to 4.0, the...
Embodiment 3
[0102] Embodiment 3, the enzymatic property determination of recombinant chitosanase BsCsn46
[0103] 1. Determination of optimum pH and pH stability
[0104] Determination of optimum pH: use the purified chitosanase solution obtained in Example 2 as the enzyme solution to be tested, and determine the optimum pH within the pH range of 3.5-7.5. Enzyme activity was determined according to the standard method in Example 2. Taking the highest value of enzyme activity as 100%, the relative enzyme activities under different pH conditions were calculated respectively.
[0105] Determination of pH stability: Dilute the enzyme solution to be tested with different buffer solutions within the pH range of 3.0-11.0, incubate at 45°C for 30 minutes, immediately cool in an ice-water bath for 30 minutes, and then determine the residual enzyme under the optimal conditions according to the standard method vitality. With the enzyme solution without the above treatment (the above treatment ref...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com