Pear transcription factor PbrMYB169 and application thereof
A technology of transcription factors and genes, applied in the fields of application, genetic engineering, plant genetic improvement, etc., can solve the problems of reducing fruit quality, achieve the effect of reducing agricultural costs and achieving environmental friendliness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
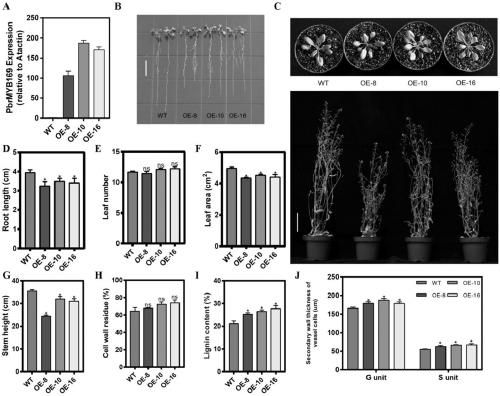
[0028] Example 1 Analysis of spatio-temporal expression pattern of PbrMYB169 gene
[0029] Different tissue samples of ‘Dangshan Suli’ were collected from Gaoyou Orchard, Jiangsu Province (2015). Pear varieties with high and low stone cell content were collected from the National Sand Pear Germplasm Resource Garden. Total RNA was extracted by CTAB method (Porebski et al., 1997), and the quality of the extracted samples was detected by spectrophotometer and agarose gel. Take 3 μg of extracted total RNA, and use one-step gDNA removal and cDNA synthesis kit (Transgen, China) for reverse transcription, and the method refers to the instruction manual. The primers used in the fluorescent quantitative PCR were gene-specific primer pairs: SEQ ID No.5 and SEQ ID No.6; GAPDH was used as an internal reference gene, and the fluorescent quantitative kit was purchased from Roche Company. The instrument used for Real-timePCR was Roche 480 quantitative PCR instrument, and the reaction syste...
Embodiment 2
[0031] Example 2 Isolation and Cloning of PbrMYB169 Gene
[0032]Take 3 μg of early pulp RNA of ‘Dangshan Suli’, and use one-step gDNA removal and cDNA synthesis kit (Transgen, China) for reverse transcription, and the method refers to the instruction manual. The amplification gene primer pair is SEQ ID No.3 and SEQ ID No.4. The 50 μL reaction system includes 200ng cDNA, 1× buffer (TransStart FastPfu Buffer), 10mM dNTP, 1U Taq polymerase (TransStart FastPfu DNA Polymerase) ( The aforementioned buffer solution and Taq polymerase were purchased from TRANS Company), 500 nM of the aforementioned primers. The PCR reaction was completed on the eppendorf amplification instrument according to the following program: 95°C, pre-denaturation for 2 minutes, denaturation at 95°C for 20 seconds, annealing at 60°C for 20 seconds, extension at 72°C for 1 minute, 35 thermal cycles, extension at 72°C for 10 minutes, Store at 4°C. Produces a single PCR band product.
[0033] After the PCR prod...
Embodiment 3
[0034] Embodiment 3 Construction of Plant Transformation Overexpression Vector
[0035] According to the multiple cloning site of the pCAMBIA-1301 vector and the analysis of the enzyme cutting sites on the coding region sequence of the PbrMYB169 gene, Xba I and BamH I were selected as endonucleases. According to the general principles of primer design, primers SEQ ID NO.3 and SEQ ID NO.4 with restriction sites were designed with Snapgene software. The annealing temperature of PCR amplification is 55° C., and the PCR reaction system and amplification procedure are the same as in Example 2. Recover the target band and connect it to the pEASY vector.
[0036] The total volume of the double enzyme digestion system was 40 μL, which contained 12 μL of pEASY-PbrMYB169 plasmid, 4 μL of 10×Buffer (purchased from NEB Company), 0.8 μL of Xba I, 0.8 μL of BamH I and 22.2 μL of water. The total volume of the pCAMBIA1301 vector double enzyme digestion system was 40 μL, which contained 8 μ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com