Biological method capable of increasing resveratrol accumulation amount
A technology for resveratrol and the highest yield, applied in the field of bioengineering, can solve the problems of lack of cofactors and low enzyme activity, and achieve the effect of increasing production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Example 1 Construction of two expression plasmids
[0052] 1.1 Primer design
[0053] According to the Rgtal, Pc4cl and Ahsts sequences reported by NCBI, the following 6 pairs of primers were designed:
[0054] Rgtal-F: 5′C CCATGG GCATGGCGCCTCGCCCGACTTCG 3′
[0055] Rgtal-R: 5′CGC GGATCC TGTGCCAGCATCTTCAGCAGAAC 3′
[0056] 4CL-STS-1F: 5′ CATATG GGCGATTGCGTGGCC 3′
[0057] 4CL-STS-1R: 5′GCCAGCGGCGATCTGCCGAAA GGAAGCGGA ATGGTTAGCGTGAGTGGTATC 3′
[0058] 4CL-STS-2F: 5′GATACCACTCACGCTAACCA TTCCGCTTCC TTTCGGCAGATCGCCGCTGGC 3′
[0059] 4CL-STS-2R: 5′ CTCGAG TTAAATGGCCATGCTAC 3′
[0060] 4CL-STS-1NF: 5′GTATAAGAAGGAGATATA CATATG GGCGATTGCGTGGCC 3′
[0061] 4CL-STS-2NR: 5′CGGTTTCTTTACCAGA CTCGAG TTAAATGGCCATGCTACGC 3′
[0062] AccBC-F: 5′ATAAGGAGATATA CCATGG GCGTGTCAGTCGAGACTAGGAAG 3′
[0063] AccBC-R: 5′GCCGAGCTCGAATTC GGATCC TGCTTGATCTCGAGGAGAAC 3′
[0064] PTDH-F: 5′ GCAGATCTCCAATTG GATATC GATGCTGCCGAAACTGGTCATC 3′
[0065] PTDH-R: 5′CTTTACCAGAC...
Embodiment 2
[0081] Example 2 Construction of fumB gene knockout strain
[0082] 2.1 Primer design
[0083] A total of 4 primers are needed to knock out fumB gene, primers S1 and S2 for homologous recombination and primers S3 and S4 for identifying defective strains.
[0084] S1: 5'ATGTCAAACAAACCCTTTATCTACCAGGCACCTTTCCCGAAGCGATTGTGTAGGCTGGAG
[0085] S2: 5'TTACTTAGTGCAGTTCGCGCACTGTTTGTTGACGATTTGCACGGCTGACATGGGAATTAG
[0086] S3: 5'CCTGCGCTATTCACAGAATG
[0087] S4: 5'ATTGCAGCAGATCCACGTAG
[0088] 2.2 Construction of gene-deficient strain ΔfumB
[0089] The homologous recombination helper plasmid pKD46 was transformed into the host strain E.coli BL21(DE3), induced by L-arabinose to prepare electroporation competent cells.
[0090]The plasmid pKD4 containing the kanamycin resistance gene kana was used as a template, and S1 and S2 were used as primers for PCR amplification. Add 2 μL of the resistant fragment pKD4-kana to electroporation competent cells on ice, mix well and place in a 1 m...
Embodiment 3
[0093] Example 3 Transformation of expression plasmids to synthesize resveratrol strains
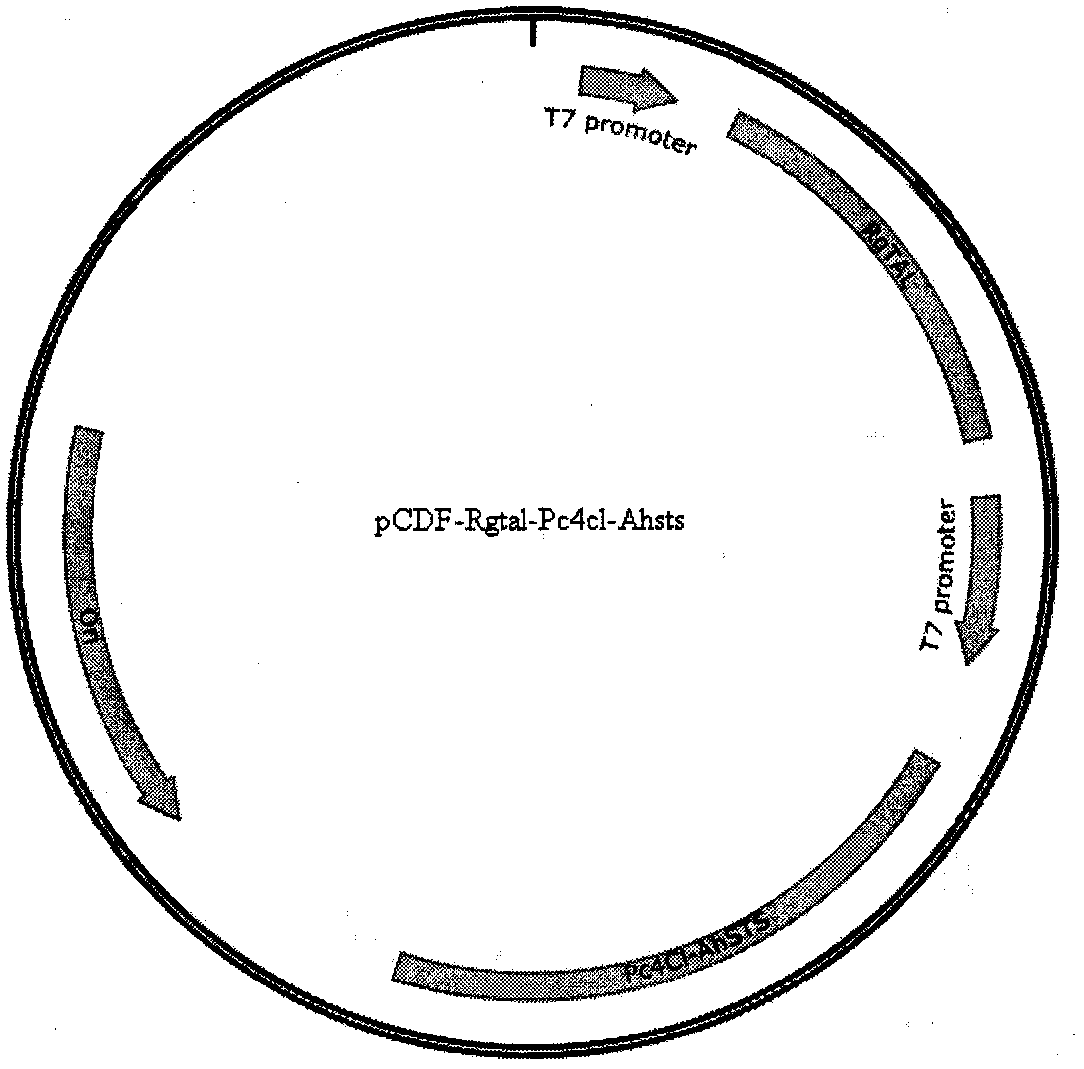
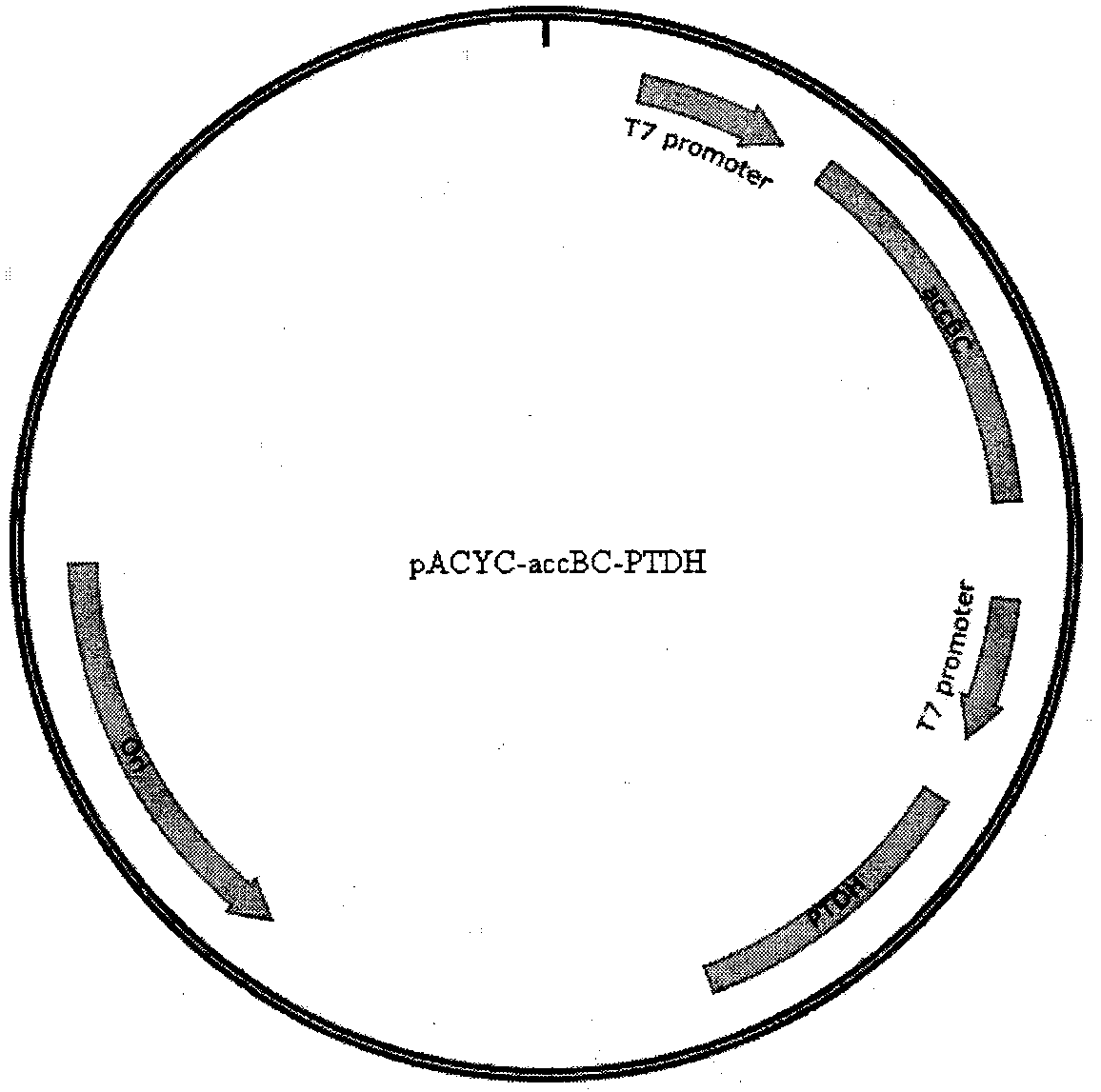
[0094] The ΔfumB gene-deficient strain was made into a ΔfumB gene-deficient E. coli competent according to the standard procedure for E. coli competent production, and the successfully constructed pCDFduet-Rgtal-Pc4cl-Ahsts and pACYCduet-AccBC-PDTH recombinant plasmids were transformed into the gene-deficient strain ΔfumB, And spread a solid LB plate containing spectinomycin and chloramphenicol in one thousandth, and cultivate at 37°C until the transformants grow out. Among them, the gene-deficient strain ΔfumB is a strain without spectinomycin and chloramphenicol resistance, and cannot grow on solid LB plates containing spectinomycin and chloramphenicol. Therefore, pCDFduet-Rgtal-Pc4cl containing pCDFduet-Rgtal-Pc4cl was obtained by screening with two antibiotics. - Gene-deficient strains of the Ahsts and pACYCduet-AccBC-PTDH recombinant plasmids. Designated as pCDF-ΔfumB-Res
[0095]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com