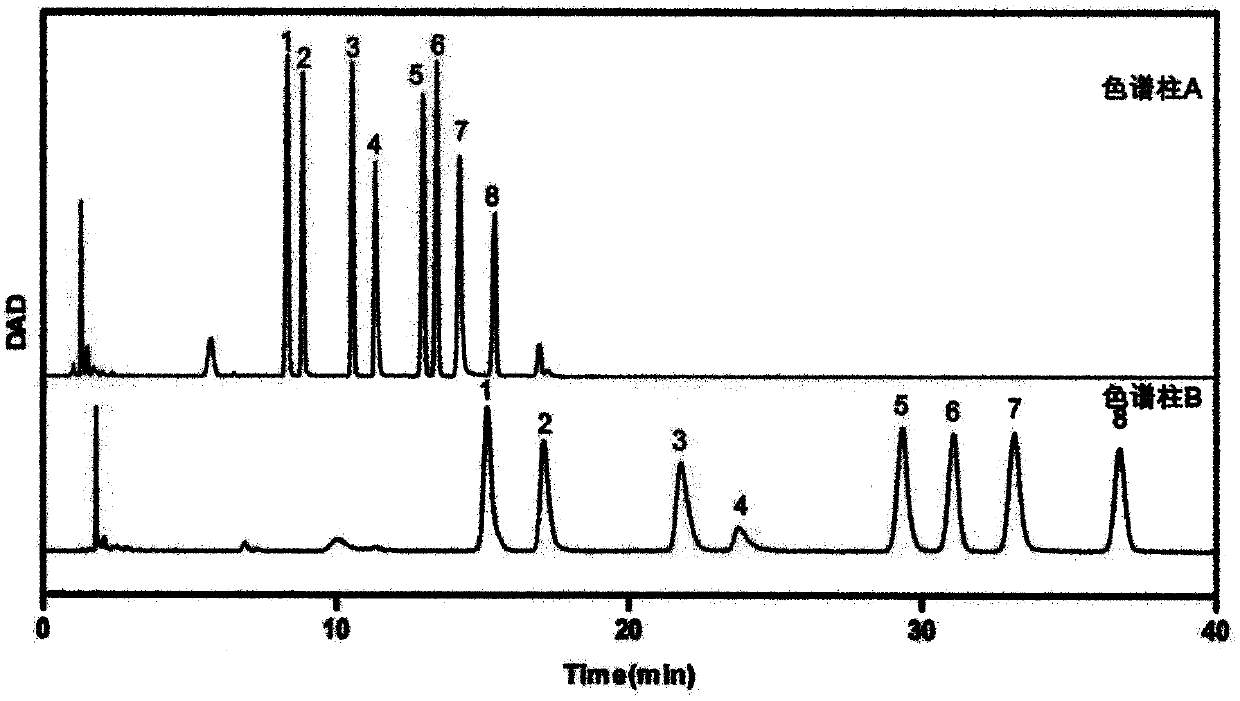
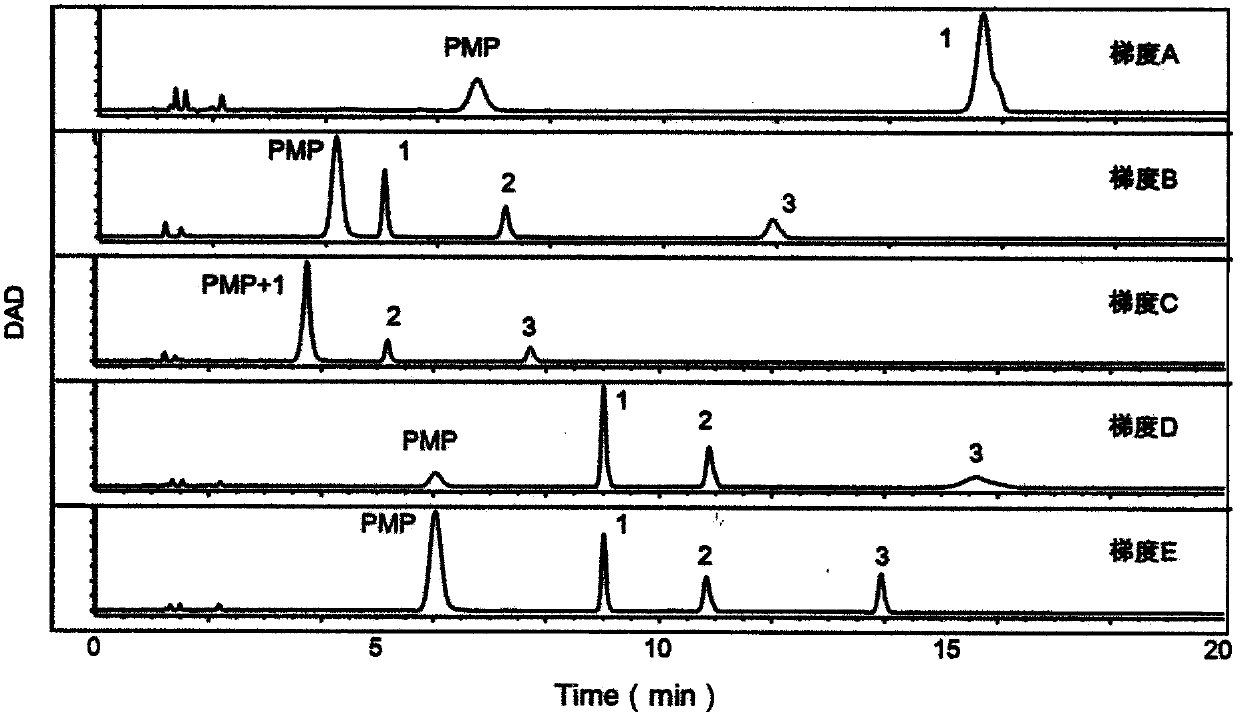
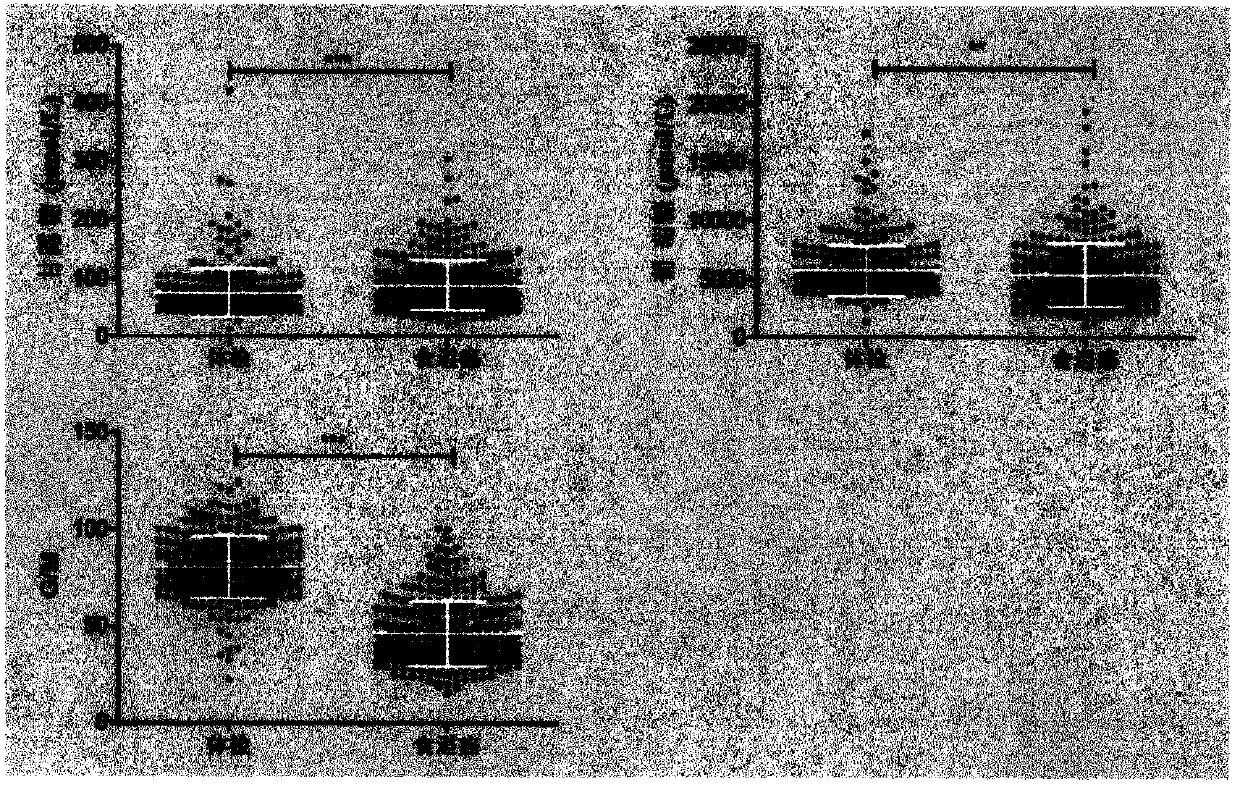
Method for identifying biomarker of esophageal cancer and detection kit thereof
A biomarker and esophageal cancer technology, applied in the field of medicine, can solve the problems of being unsuitable for routine purposes, large volume of serum samples, and expensive instruments, and achieve the effects of simplified operation, short analysis time, and reasonable instrument prices
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] (1) Precisely weigh mannose (Man), glucosamine (GlcN), galactosamine (GalN), glucuronic acid (GlcUA), glucose (Glc), galactose (Gal), xylose (Xyl), rock Proper amount of fucose (Fuc), add deionized water to prepare two mixed standard solutions containing the above monosaccharides 0.1mg / mL, ready to use;
[0053] (2) Create an alkaline environment: Take 40μL of mixed monosaccharide standard in a 1.5mL EP tube, add 40μL of 0.3mol / L sodium hydroxide, vortex to mix;
[0054] (3) PMP derivatization: add 60μL 0.5mol / L 1-phenyl-5-methylpyrazolone (PMP) to each sample, vortex to mix, centrifuge and react in an oven at 70°C for 1h;
[0055] (4) Acid neutralization reaction: take out the samples in the oven, leave them to cool, add 40μL 0.3mol / L hydrochloric acid to each sample, vortex and mix;
[0056] (5) Extraction: add 500μL of chloroform to each tube, vortex, centrifuge, remove the lower layer of chloroform, save the supernatant, repeat 3 times;
[0057] (6) Centrifuge the sample at ...
Embodiment 2
[0080] (1) Precisely weigh appropriate amounts of mannose (Man), rhamnose (Rha) and glucose (Glc), and add deionized water to prepare the same 5 portions of the same 0.1 mg / mL mixed standard solution containing the above monosaccharides. Match
[0081] (2) Create an alkaline environment: Take 40μL of mixed monosaccharide standard in a 1.5mL EP tube, add 40μL of 0.3mol / L sodium hydroxide, vortex to mix;
[0082] (3) PMP derivation: add 60μL 0.5mol / L PMP to each sample, vortex to mix, centrifuge and react in an oven at 70°C for 1h;
[0083] (4) Acid neutralization reaction: take out the samples in the oven, leave them to cool, add 40μL 0.3mol / L hydrochloric acid to each sample, vortex and mix;
[0084] (5) Extraction: add 500μL of chloroform to each tube, vortex, centrifuge, remove the lower layer of chloroform, save the supernatant, repeat 3 times;
[0085] (6) Centrifuge the sample at 13000r / min for 10min, take 80μL of supernatant into a brown sample bottle equipped with a 150μL inner ...
Embodiment 3
[0106] (1) Precisely weigh an appropriate amount of mannose and glucose, add deionized water to prepare the above monosaccharides 0.5mg / mL, 0.25mg / mL, 0.1mg / mL, 0.05mg / mL, 0.01mg / mL, 0.005mg / mL, 0.0025mg / mL, 0.001mg / mL, 0.0005mg / mL mixed standard solution, ready to use;
[0107] (2) Create an alkaline environment: Take 40μL of mixed monosaccharide standard in a 1.5mL EP tube, add 40μL of 0.3mol / L sodium hydroxide, vortex to mix;
[0108] (3) PMP derivation: add 60μL 0.5mol / L PMP to each sample, vortex to mix, centrifuge and react in an oven at 70°C for 1h;
[0109] (4) Acid neutralization reaction: take out the samples in the oven, leave them to cool, add 40μL 0.3mol / L hydrochloric acid to each sample, vortex and mix;
[0110] (5) Extraction: add 500μL of chloroform to each tube, vortex, centrifuge, remove the lower layer of chloroform, save the supernatant, repeat 3 times;
[0111] (6) Centrifuge the sample at 13000 r / min for 10 min, take 80 μL of supernatant into a brown sample bott...
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com