Omega-transaminase from bacillus pumilus and application in biological amination
A transaminase, amino technology, applied in the field of bioengineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Example 1 Obtaining of Bacillus pumilus ω-transaminase gene
[0052] Firstly, the whole gene DNA of Bacillus pumilus W3 was extracted, and specific primers were designed according to the ω-transaminase gene sequence.
[0053] Using the whole gene DNA of Bacillus pumilus W3 as a template, and using FWD-ota and REW-ota as primers for PCR, a 915bp product fragment was obtained. After the product was sequenced and compared with NCBI, it was found that it was related to the Bacillus pumilus W3ω-transaminase gene The sequence of (Sequence ID: MH196528) is consistent, and the sequence ID is expected to be published on NCBI in May 2019.
[0054] Wherein, design primer FWD-ota (sequence as shown in SEQ ID NO.4) and REW-ota (sequence as shown in SEQ ID NO.5), utilize PCR to the DNA coding frame 5' and 3' of Bacillus pumilus W3ω-transaminase XhoI and PstI restriction enzyme sites (without signal peptide) are respectively introduced on both sides, the underline is the restriction ...
Embodiment 2
[0070] The predicted crystal structure of Bacillus pumilus W3ω-transaminase and its amino acid similarity with Arthrobacter sp.KNK 168ω-transaminase were obtained by "homology modeling".
[0071] The full length of the Bacillus pumilus ω-transaminase gene ota3 is 915bp, and the predicted open reading frame of the new protein is located at 30-915 nucleotides, encoding 304 amino acid residues, and the molecular weight is 33.4kDa.
[0072] Submit the amino acid sequence of Bacillus pumilus ω-transaminase to the SWISS-MODEL protein online modeling server (http: / / swissmodel.expasy.org / ) for homology modeling, and then use Discovery studio software to analyze the homology of Bacillus pumilus ω-transaminase protein Modeling structural analysis (see figure 2 ): The template for homology modeling of this protein is 5e25.1.A, and the sequence homology similarity with the template is 51.21%.
[0073] The ω-transaminase gene sequence of Bacillus pumilus obtained from the screened Bacill...
Embodiment 3
[0074] Example 3 Construction of Bacillus pumilus ω-transaminase prokaryotic expression vector, recombinant expression and protein expression thereof
[0075] 1. Construction of prokaryotic expression vector
[0076] (1) Primer design: design primers from the mature peptide sequence after the signal peptide (as shown in SEQ ID NO: 4 and SEQ ID NO: 5)
[0077] (2) PCR reaction, using the cloning vector pMD-19T-ota3 as a template, annealing at 62°C, 35 cycles.
[0078] (3) The PCR product of ota3 and the plasmid pColdⅡ were double-digested with XhoI and PstI.
[0079] Table 3 Double Enzyme Digestion System
[0080] Element
Usage amount
Purification of PCR products / plasmids
30μl
10*quitcut buffer
5μl
QuitCut SnaBI
1μl
Quit Cut Not I
1μl
wxya 2 o
13μl
total capacity
50μl
[0081] Digest at 37℃ for 2hr
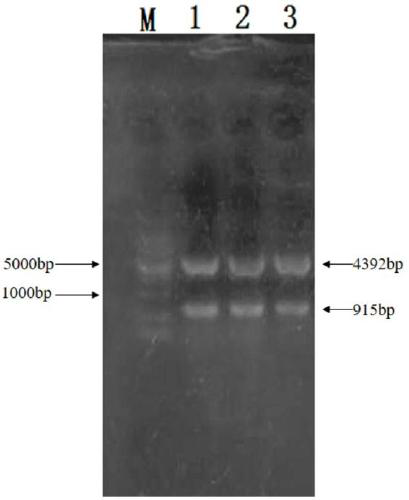
[0082] (4) Connect, transform, and carry out double-enzyme digestion identification (see ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com