PCR rapid detection method for marine culturable bacteria, and kit
A technology of marine bacteria and kits, applied in the field of microbial detection, to achieve the effects of increasing yield, shortening extraction time, and increasing specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
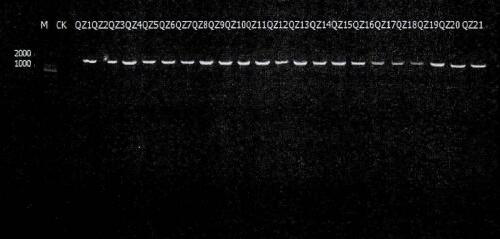
[0045] PCR rapid test of cultivable bacteria in South China Sea sediments
[0046] S1. Sample processing
[0047] Put 10g of the marine sediment sample taken from the South China Sea into 80ml sterile seawater with glass beads, incubate at 28℃, 140rpm shaker for 30min, then transfer 1ml suspension into 9ml sterile seawater and mix well. 10 -1 -10 -5 Five different concentration gradients. Using the spreading plate method, 0.1ml samples with different concentration gradients were spread on the entire solid medium, four in parallel, two by one placed in an incubator at 28°C and 10°C for 2-7 days. Purification is performed according to the morphological characteristics of the colony after culture.
[0048] extract
[0049] Pipette 100 µL of 10% (w / v) sterile Chelex-100 solution into a 1.5mL Eppendorf tube, use a sterilized toothpick to pick out the purely cultured colonies, pick the size of the rice grains to the tube wall, crush and disperse Dissolve and mix the Chelex-100 solution. ...
Embodiment 2
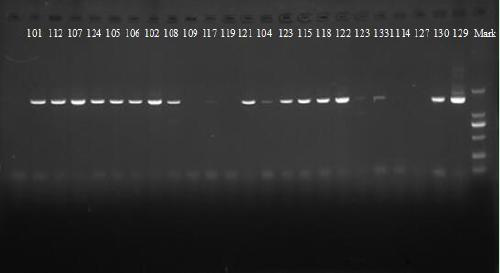
[0058] Rapid PCR Test of Symbiotic Actinomyces from Gorgonian Coral in the North Sea
[0059] S1. Sample processing
[0060] Gorgonians to be collected in the North Sea Anthogorgia caerulea Take 10g of the sample and grind it with a mortar and put it into a 100ml triangular bottle with sterile seawater, take it out after 2 days in a shaker at 28°C and 140 rpm, and take the supernatant to prepare 10 -1 -10 -5 At different dilutions, draw 0.5 ml of each multiple of dilutions and spread on the solid culture, and incubate at 28°C for 3-5 days. After being cultured, the colonies are first distinguished by naked eyes according to the characteristics of the colonies, marked, and then further observed with a microscope to determine the separation object, and then a single colony is picked with a sterilized bamboo stick, and the quartering method is repeated for separation and purification.
[0061] DNA extraction of symbiotic actinomycetes
[0062] After the purified strain, a certain amount...
Embodiment 3
[0071] A kit for rapid PCR of marine cultivable bacteria, which is characterized by comprising: marine bacterial DNA extraction reagents, PCR reaction reagents, and electrophoresis reagents, wherein:
[0072] (1) Marine bacterial DNA extraction sample preparation and extraction reagent concentration: colonies of marine cultivable bacteria purely cultured in seawater medium, 10% (w / v) sterile Chelex-100 solution;
[0073] (2) Each raw material and its concentration in the PCR reaction reagent: 16S rDNA universal primer (27F, 1492R), 10×PCRBuffe, 5U / Taq enzyme, 10Mm dNTP and ddH 2 O; Among them, the universal primer sequences are 27F and 1492R in the prior art, and their sequences are:
[0074] 27F: 5ʹ-AGAGTTTGATCC TGGCTCAG-3ʹ;
[0075] 1492R: 5ʹ-GGTTACCTTGTTACGA CTT-3ʹ.
[0076] (3) Electrophoresis reagents: agarose, 50×TBE, Goldview II nucleic acid dye, DNA Marker;
[0077] A method for rapid PCR with a kit for rapid PCR detection of marine cultivable bacteria is characterized in that it...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com