Preparation method and application of targeting exosome
An exosome, targeting technology, applied in the field of biomedicine, can solve the problem of low exosome modification efficiency, and achieve the problem of low transfection efficiency, difficulty in large-scale production, simple modification means, and good cell targeting. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0048] The present invention provides a method for preparing targeted exosomes. After the PSP1 polypeptide is covalently combined with any targeting peptide or a short peptide with immune function to form a PSP1 targeting peptide, it is then combined with exosomes , that is, to obtain targeted exosomes; wherein, the amino acid sequence of the PSP1 polypeptide is: CLSYYPSYC, or a polypeptide having a sequence of more than 80% homology with PSP1.
[0049] Now, EGFR targeting peptide (GE11), nAchR targeting peptide (RVG29), liver cancer cell targeting peptide (SP94) and PSP1 polypeptide are combined to form PSP1 targeting peptide: PSP1-GE11, PSP1-RVG29-6His and PSP1-SP94 , and then modify the surface of exosomes as Example 1, Example 2 and Example 3; of course, the present invention describes not only the above targeting peptides, but all targeting peptides under the concept of the present invention or have Short peptides that promote immune function, that is, the above examples ...
Embodiment 1
[0058] Example 1: Preparation of PSP1-GE11
[0059] GE11 is a polypeptide with 12 amino acid residues (amino acid sequence: YHWYGYTPQNVI) with EGFR targeting, and is not derived from EGF, and has no EGFR activation activity.
[0060] Using chemical synthesis method, the amino acid sequence is directly synthesized: CLSYYPSYCYHWYGYTPQNVI 21 amino acid residue polypeptide with bidirectional binding ability of phosphatidylserine and EGFR, that is, PSP1-GE11 is obtained.
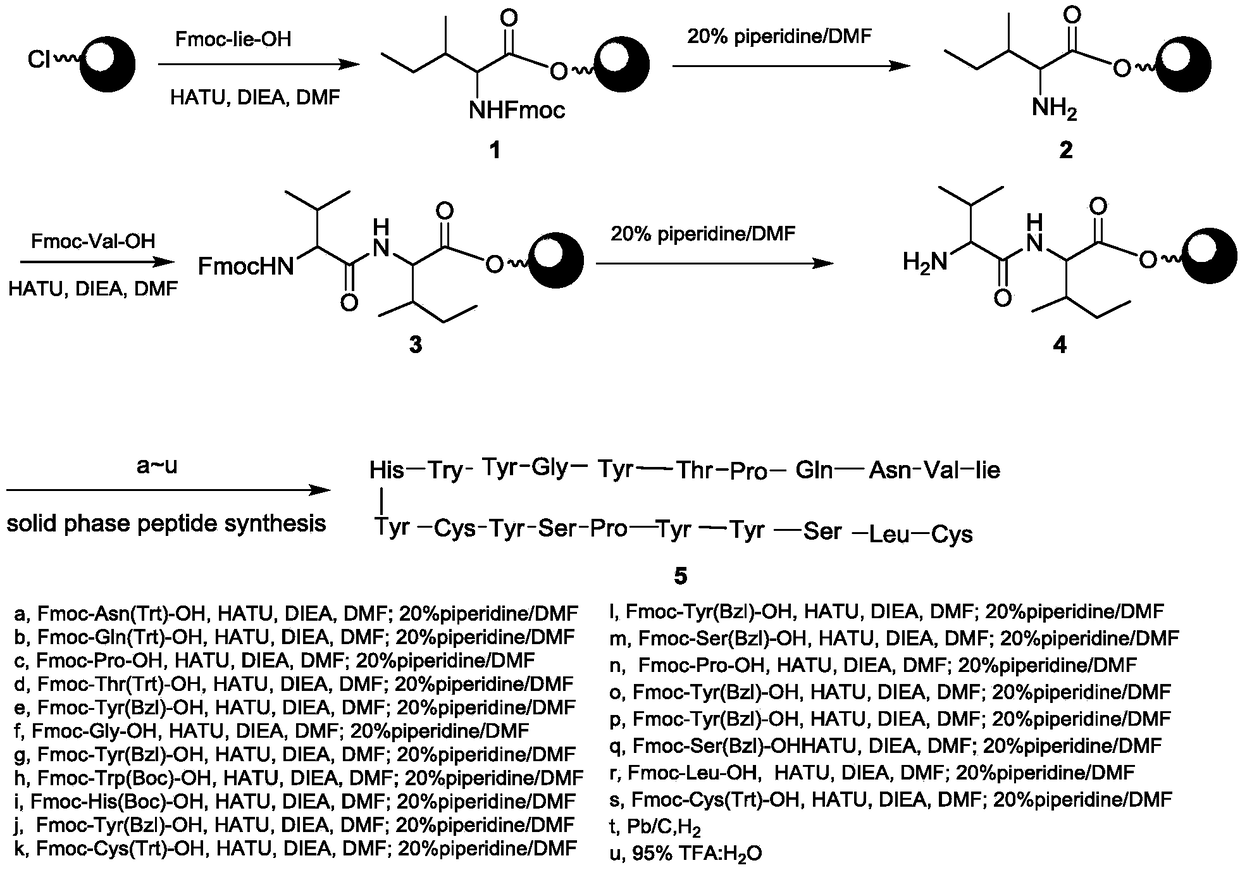
[0061] Wherein, the chemical synthesis method can adopt any synthetic method in the field of chemistry or biology. The present invention provides the Fmoc solid-phase polypeptide synthesis method as one of the examples, using 2-Cl-Trt (2-chlorotrityl chloride ) resin as a solid phase carrier. Its synthetic route is as follows figure 1 shown; the specific steps are as follows:
[0062] 1. Add a certain amount of 2-Cl-Trt resin (1eq) and an appropriate amount of DIEA (N, N diisopropylethylamine, 5eq) to an appro...
Embodiment 2
[0066] Example 2: Preparation of PSP1-RVG29-6His
[0067] Neurotropic virus-derived peptide (RVG29) (amino acid sequence: YTIWMPENPRGTPCDIFTNSRGKRASNG) is derived from the RVG protein on the capsid of rabies virus, and can be specifically recognized by the nicotinic acetylcholine receptor (nAchR) on the surface of nerve cells.
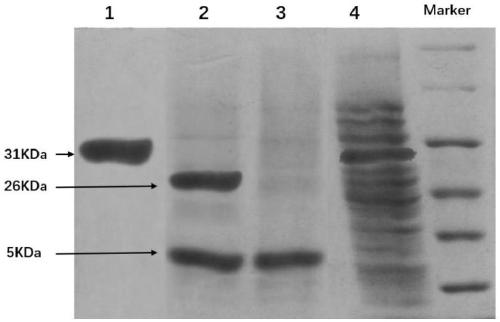
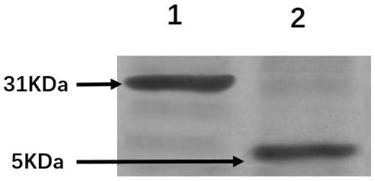
[0068] By means of biological expression, the prokaryotic expression system of PSP1-RVG29-6His (CLSYYPSYCYTIWMPENPRPGTPCDIFTNSRGKRASNGHHHHHH) with 6 histidine residues was constructed, and the specific steps are as follows:
[0069] Step a, synthesize 147 pairs of DNA sequences each containing BamHI and XhoI restriction endonuclease sites at the 5' end and the 3' end: 5'-ggatccTGCCTGTCCTATTATCCGTCCTATTGCTACACTATTTGGATGCCGGAAAATCCGCGTCCGGGTACCCCATGCGACATCTTCACCAACTCCGTGGTAAACGCGCGTCTAACGGCCACCATCACCACCACCATTAActcgag-3';
[0070]Step b, 147 pairs of DNA sequences of PSP1-RVG29-6His and pGEX-4T-1 plasmid were double digested with BamHI and XhoI restrictio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com