Detection method of viable bacterium number of embedded probiotic product
A detection method and technology of probiotics, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve problems such as large detection errors and unscientific results, achieve small counting errors, reliable detection results, and improve stability Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
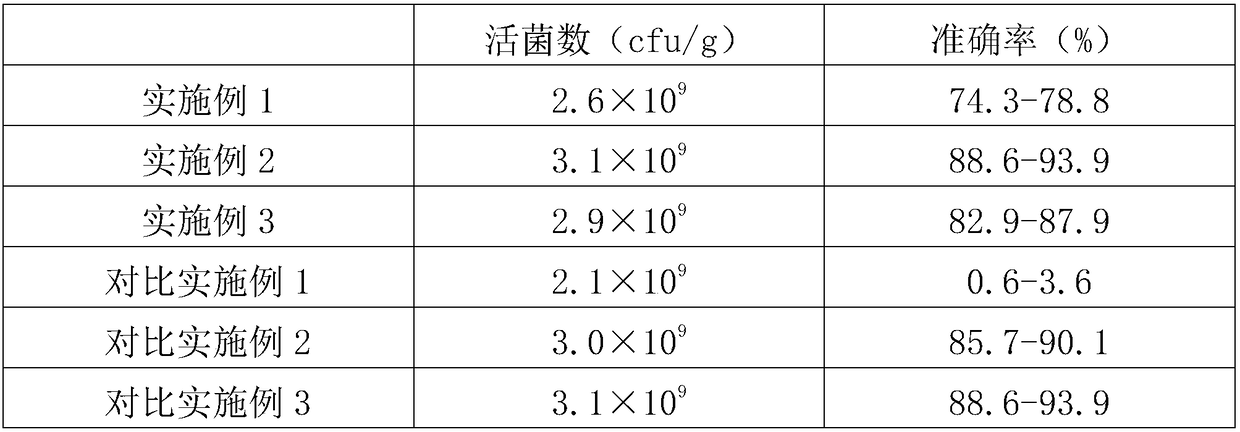
[0029] A method for detecting the number of viable bacteria in an embedded probiotic product, comprising the following steps:
[0030] (1) Pretreatment: Weigh an appropriate amount of the embedded probiotic product to be tested and grind it into a fine powder in a sterile mortar;
[0031] (2) Preparation of bacterial suspension: Weigh 1 mg of the fine powder prepared in step (1), add it to a 50ml centrifuge tube, add 30ml of dilution buffer, vortex shaker to fully dissolve the embedding layer, and obtain bacterial suspension liquid;
[0032] (3) Gradient dilution: oscillate and mix the bacterial suspension prepared in step (2), take 1ml of the mixed bacterial suspension, add dilution buffer to dilute into 4 concentration gradients, and the corresponding dilution factor is 5 times , 50 times, 500 times, 5000 times;
[0033] (4) Bacterial colony culture: draw the gradient dilution in step (3) and add in the sterile petri dish, add MRS culture medium and mix, leave standstill, ...
Embodiment 2
[0044] A method for detecting the number of viable bacteria in an embedded probiotic product, comprising the following steps:
[0045] (1) Pretreatment: Weigh an appropriate amount of the embedded probiotic product to be tested and grind it into a fine powder in a sterile mortar;
[0046] (2) Preparation of bacterial suspension: Weigh 3mg of the fine powder prepared in step (1), add it to a 50ml centrifuge tube, add 40ml of dilution buffer, vortex shaker to fully dissolve the embedding layer, and obtain bacterial suspension liquid;
[0047] (3) Gradient dilution: Shake and mix the bacterial suspension prepared in step (2), take 3ml of the mixed bacterial suspension, add dilution buffer to dilute into 4 concentration gradients, and the corresponding dilution factor is 10 times , 100 times, 1000 times, 10000 times;
[0048] (4) Bacterial colony culture: draw the gradient dilution in step (3) and add in the sterile petri dish, add MRS culture medium and mix, leave standstill, a...
Embodiment 3
[0059] A method for detecting the number of viable bacteria in an embedded probiotic product, comprising the following steps:
[0060] (1) Pretreatment: Weigh an appropriate amount of the embedded probiotic product to be tested and grind it into a fine powder in a sterile mortar;
[0061] (2) Preparation of bacterial cell suspension: Weigh 5 mg of the fine powder prepared in step (1), add it to a 50 ml centrifuge tube, add 50 ml of dilution buffer, vortex shaker to fully dissolve the embedding layer, and obtain the bacterial cell suspension liquid;
[0062] (3) Gradient dilution: oscillate and mix the bacterial suspension obtained in step (2), take 5ml of the mixed bacterial suspension, add dilution buffer to dilute into 4 concentration gradients, and the corresponding dilution factor is 15 times , 150 times, 1500 times, 15000 times;
[0063] (4) Bacterial colony culture: draw the gradient dilution in step (3) and add in the sterile petri dish, add MRS culture medium and mix...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com