Whole-bacterium enzymic preparation for catalyzing saccharification of lignocellulose
A technology of lignocellulose and whole-bacteria enzymes, applied in the biological field, can solve the problems of high cost and cumbersome process, and achieve the effects of improving synergy, reducing consumption, maintaining stability and activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1: Construction of Clostridium thermocellum expressing exocrine β-1,4 glucosidase
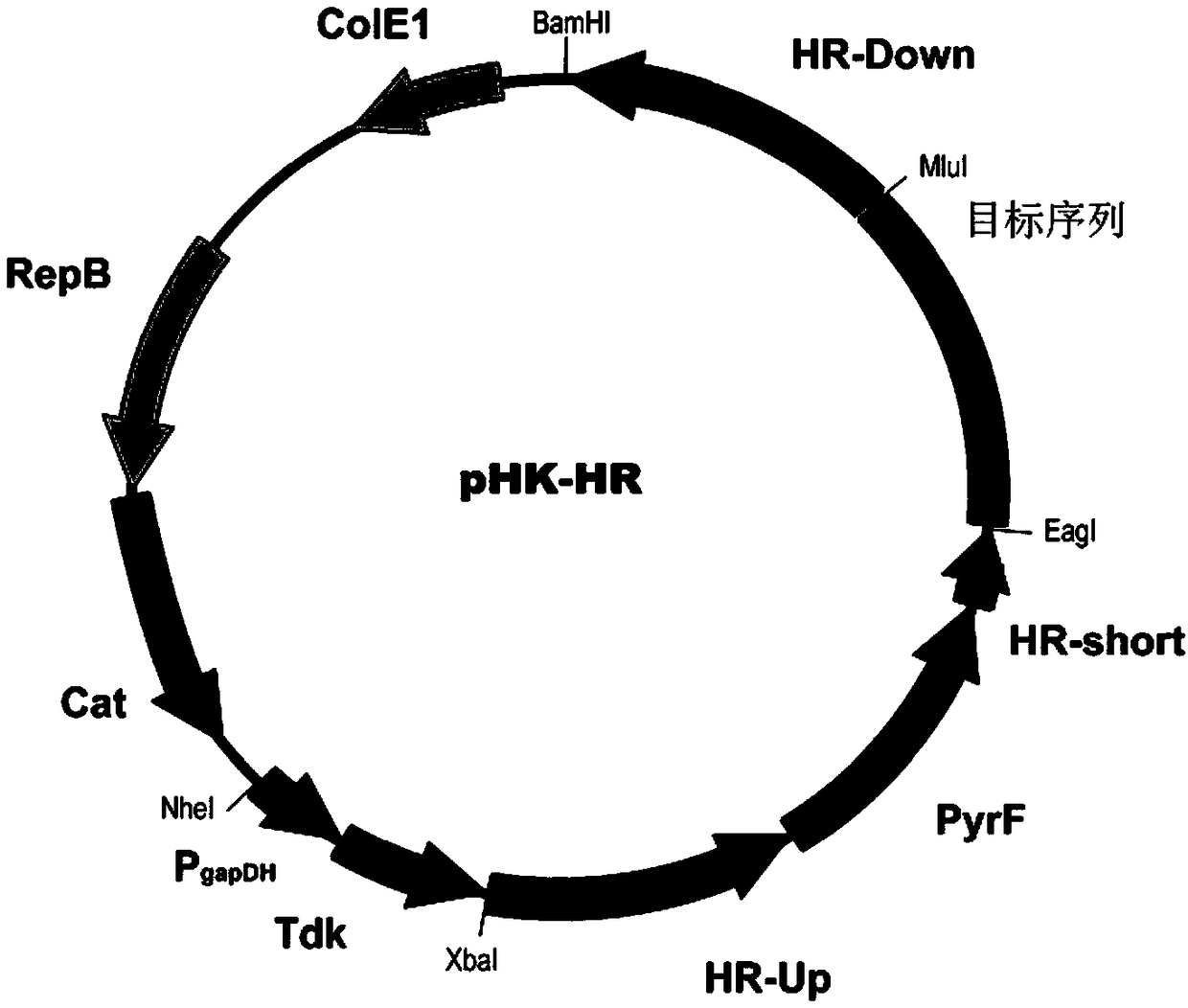
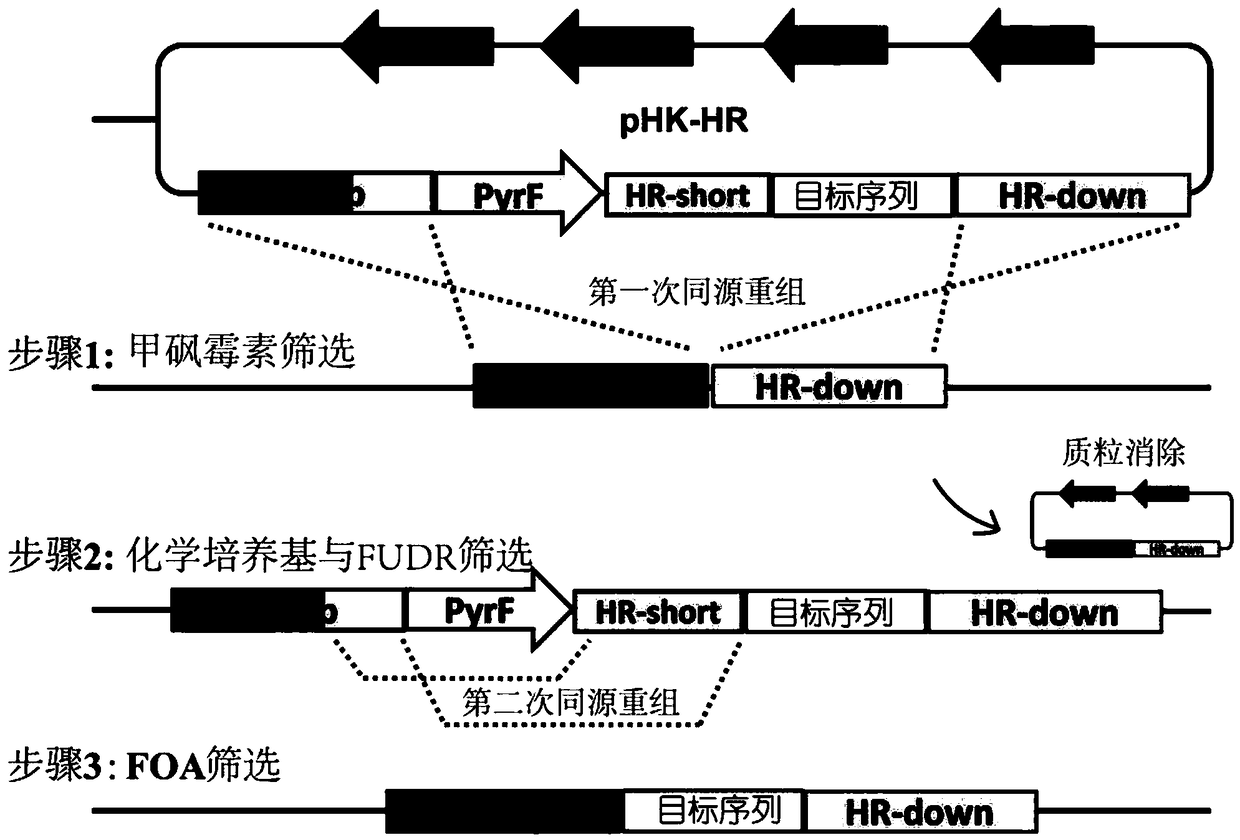
[0040] The cellulase Cel9K (exocellulase, encoded by nucleotide sequences 2113813 to 2111293 in genome CP002416.1) in the cellulosome of Clostridium thermocellum was selected as a targeted knock-in site between the enzymatic catalytic domain and the docking module point. First, the β-1,4-glucosidase BglA (Genbank sequence number is AFO70070.1) encoding gene was used as the target sequence, and the restriction sites of MluI and EagI were used to clone into the homologous recombination plasmid pHK-HR ( figure 2 ), the homologous recombination plasmid pHK-HR-BglA was constructed. The HR-up sequence of the upstream homology arm is the nucleic acid sequence of 2111347 to 2112870 in the genome of Clostridium thermocellum DSM1313 (the sequence number in the NCBI database is CP002416.1), and the HR-down of the downstream homology arm is the nucleic acid sequence of 2109848 to 2111354 i...
Embodiment 2
[0045] Example 2: Construction of a whole bacterial enzyme preparation based on Clostridium thermocellum cellosomes by direct linking
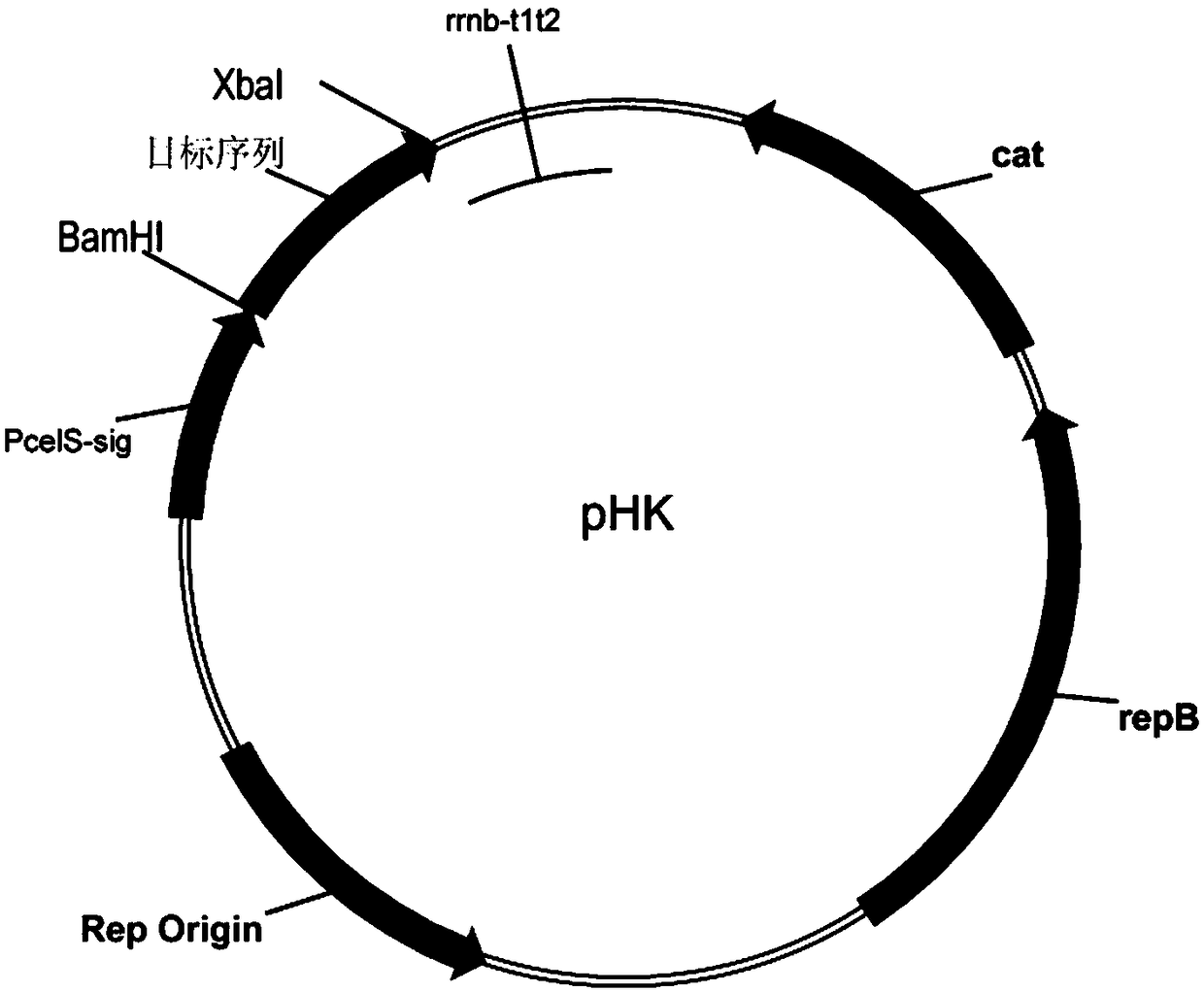
[0046] Using the method of overlap extension polymerase chain reaction, xylanase XynC (SEQ ID NO: 1) was combined with the sequence of the type II adhesion module CohIIct (SEQ ID NO: 10) or the type I docking module DocIct of Clostridium thermocellum The sequences of XynC (SEQ ID NO: 9) were directly linked, wherein the sequence of CohIIct or DocIct was linked to the 3' end of the XynC sequence, thereby obtaining the XynC-CohIIct and XynC-DocIct sequences. Reuse the BamHI and XbaI restriction sites, and clone the connected recombinant sequence as the target sequence into the expression plasmid pHK ( figure 1 )superior. Since pHK carries the promoter and signal peptide sequence of cellulase Cel48S derived from Clostridium thermocellum (SEQ ID NO: 11), the expressed target gene can be excreted outside the cell. The constructed plasmid was tran...
Embodiment 3
[0047] Example 3: Construction of a whole bacterial enzyme preparation based on Clostridium thermocellum cellosomes by direct linking
[0048] The xylanase XynB (SEQ ID NO: 2) encoding gene was used as the target sequence, and the cellulosic exonuclease Cel48S (encoded by the 3228088 to 3230229 nucleic acid sequences in the genome CP002416.1) of the Clostridium thermocellum cellosome was selected. The ' end was used as a targeted knock-in site, and the restriction sites of MluI and EagI were used to clone into the homologous recombination plasmid pHK-HR ( figure 2 ), the homologous recombination plasmid pHK-HR-xynB was constructed. The upstream homology arm HR-up is the nucleic acid sequence from 3230200 to 3230700 in the genome of Clostridium thermocellum DSM1313 (the sequence number in the NCBI database is CP002416.1), the downstream homology arm HR-down is the nucleic acid sequence from 3229699 to 3230199 in the DSM1313 genome, and the middle The homology arm HR-short is ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com