Method for detecting cowpat and detection kit
A kit and cow dung technology, which is applied in the field of biotechnology detection, can solve the problems of methods and inadequacies in the detection of cow feces pollution, and achieve the effects of high sensitivity, strong specificity, and strong stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1 Primer Design
[0056] 224 animal feces samples (chicken, duck, human, dog, pig, cow) were collected, and 984,224 bacterial 16SrDNA sequences were obtained by high-throughput sequencing. By comparing the obtained sequences to the database, the 16S rDNA gene sequences of Faecalibacterium in all feces samples were selected, and detailed comparisons were made between species to obtain the specific sequence of Faecalibacterium bovine. Therefore, it is speculated that the bovine Faecalibacterium can be used as a tracer microorganism to detect and distinguish cow feces, and this will be verified in follow-up experiments.
[0057] Two pairs of DNA primers CFY-1 and CFY-2 were designed for the specific sequence of bovine Faecalibacterium. The primer sequences are shown in Table 1, and FUP is the general primer for Faecalibacterium.
[0058] Table 1 Primer Sequence
[0059]
[0060] The above primers were synthesized, and the primer synthesis was completed by Meij...
Embodiment 2
[0061] Embodiment 2 specific detection
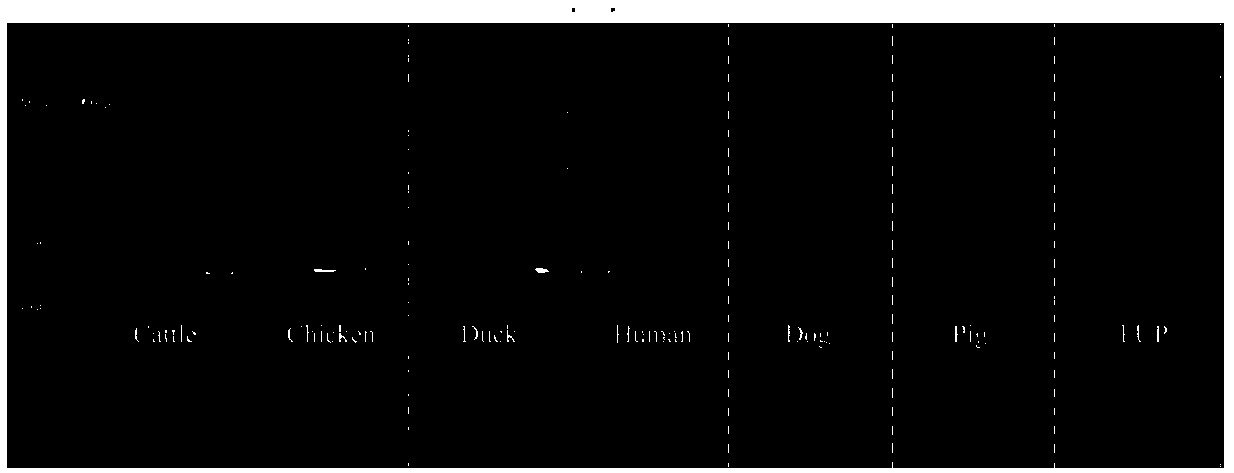
[0062] The fecal samples of six species of chicken, duck, human, dog, pig, and cow are tested, and the specific detection steps are as follows:
[0063] 1) Extract the sample genomic DNA:
[0064] A total of 224 feces samples from six species of chicken, duck, human, dog, pig, and cow were collected using MO-BIO DNAIsolation kit extracts genomic DNA from samples;
[0065] 2) Specificity verification:
[0066] Take the genomic DNA in step 1), mix the DNA samples of the same species, use CFY-1, CFY-2, FUP (universal primer) as primers respectively, add Taq PCR Master Mix (2×) 7.5 microliters, positive Add 0.5 microliters of primers, 0.5 microliters of reverse primers, and 1 microliter of genomic DNA; and add sterile water to 15 microliters; PCR reaction, the reaction temperature is as follows:
[0067] ①CFY-1: Pre-denaturation at 96°C / 300s, 1 cycle; 96°C / 30s, 54°C / 30s, 32 cycles.
[0068] ②CFY-2: Pre-denaturation at 96°C / 300s, 1 cyc...
Embodiment 3
[0073] Embodiment 3 minimum detection limit
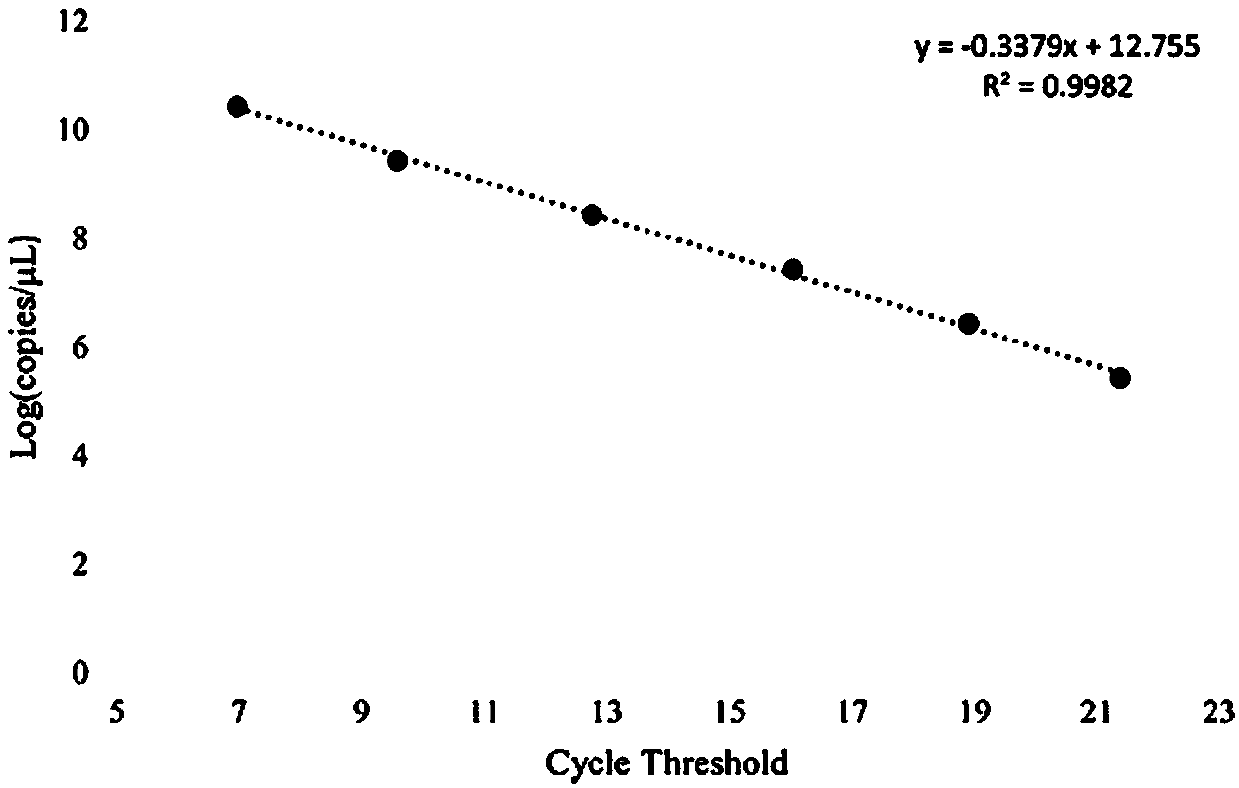
[0074] 1) draw quantitative PCR standard curve
[0075] ① Amplification of the target fragment: Use CFY-1 as a primer to amplify using ordinary PCR, and the amplification system is 100 μl.
[0076] ② Purification of PCR products: Configure 1% agarose gel electrophoresis, add 100ul PCR products into a large sample well, after the electrophoresis is completed, cut the gel under UV observation to recover the electrophoresis products, and use the recovery kit to purify the PCR products.
[0077] ③ Ligation and transformation of the target fragment with the vector: connect the target fragment with pMD 19-T vector, and pour it into DH5α Escherichia coli competent cells.
[0078] ④ Screening of recombinant plasmids: Escherichia coli in step ④ of blue-white screening, select white single colony liquid medium for expansion and culture.
[0079] ⑤ Extraction of recombinant plasmids: Use a plasmid extraction kit to extract recombinant plasm...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com