Production method, purification method and applications of protein glutaminase (PG)
A glutaminase and protein technology, which is applied in the field of screening protein glutaminase producing bacteria, can solve the problems of poor substrate specificity, few strains, low enzyme activity, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Example 1 Screening and isolation of Chryseobacterium proteolyticum (Chryseobacterium proteolyticum) YF810 bacterial strain
[0057] The YF810 strain was obtained by screening the enrichment medium, which was prepared by mixing 54mL solution A, 6mL 1% CBZ, 60μL mother solution I, 60μL mother solution II, 60μL mother solution III and 120μL mother solution IV.
[0058] Among them, the formula of liquid A is: 5g / L glucose, 0.2g / L KH 2 PO 4 , 0.2g / L MgSO 4 ·7H 2 O, 0.01% NaCl
[0059] The formula of mother liquor Ⅰ is: 20g / L CaCl 2
[0060] The formula of mother liquor Ⅱ is: 2g / L FeSO 4 ·7H 2 O, 5g / L MnSO 4 4H 2 o
[0061] The formula of mother liquor Ⅲ is: 5g / L NaWO 4 4H 2 O, 5g / L NaMO 4 2H 2 o
[0062] The formula of mother liquor IV is: 50g / L CuSO 4 ·5H 2 o
[0063] The screening steps are:
[0064] The first step is to weigh 10g of soil sample, add 90mL of sterile water, put a small amount of glass beads into an Erlenmeyer flask, and vibrate on a shaker...
Embodiment 2
[0070] Example 2 Identification of Chryseobacterium proteolyticum (Chryseobacterium proteolyticum) YF810 strain
[0071] 1. Morphological identification
[0072] (1) Observe the shape of the colony
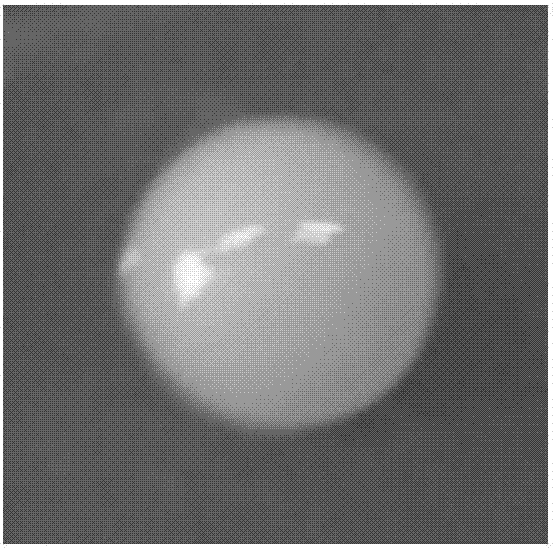
[0073] After the suspected strains were cultured on LB plates, the colonies were round, with a diameter of 2mm-4mm, neat edges, golden or orange, opaque, and the surface of the colonies was moist, smooth and shiny, such as figure 1 shown.
[0074] (2) Microscope observation
[0075] After Gram staining and spore staining, observation under an ordinary light microscope showed that the strain was rod-shaped, immobile, non-spore-free, and Gram-negative, such as figure 2 shown.
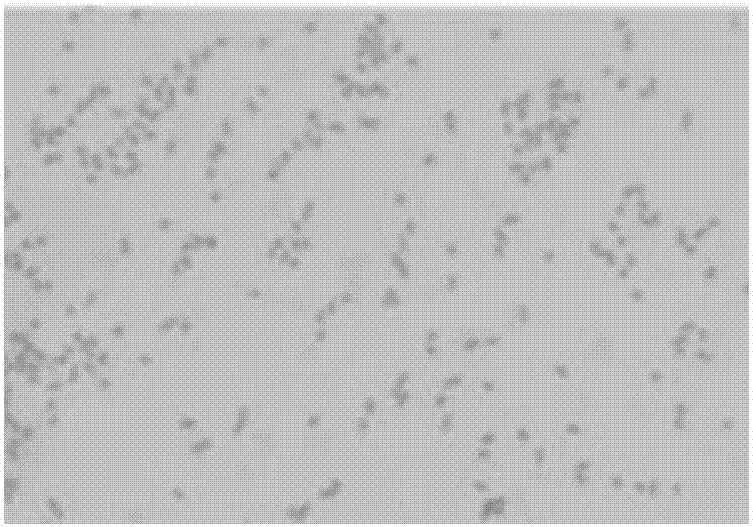
[0076] Observed under the scanning electron microscope, the bacteria are smooth without flagella, immobile, 0.2μm-0.4μm wide, 0.8μm-2.2μm long, such as image 3 shown.
[0077] 2. Physiological and biochemical identification
[0078] Physiological and biochemical identification of the Chryseobacterium...
Embodiment 3
[0090] Example 3 DNA Sequence Determination and Amino Acid Sequence Prediction of Encoding Protein Glutaminase
[0091] 1. Determination of the full-length DNA sequence of the coding protein glutaminase and its amino acid sequence prediction
[0092] Extract the total genomic DNA of the YF810 bacterial strain, and use this as a template to amplify the full-length DNA sequence of the protein glutaminase produced by the bacterial strain. The primer sequence is:
[0093] Seq ID No.4, F: 5'-CCAACCAACTTAACAAAAACTCACCATTAAAC-3'
[0094] Seq ID No.5, R: 5'-GGAACCCGAACTACCGGAGCAGGATG-3'
[0095] After the PCR product was detected by 1% agarose gel electrophoresis, it was recovered by tapping the gel and sent to a commercial company for sequencing. The sequencing work was completed by Shanghai Sangon Bioengineering Technology Service Co., Ltd.
[0096] The DNA sequence measured above, namely Seq ID No.10; was translated with the software Clone Manager to obtain the corresponding ami...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com