Novel recombination system in pseudomonas and application thereof
A recombination system, Pseudomonas technology, applied in the field of microbial genetic engineering, can solve the problems of restricting the research and transformation of Pseudomonas genome, low efficiency, and low efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0053] Preparation of electroporated cells for recombination:
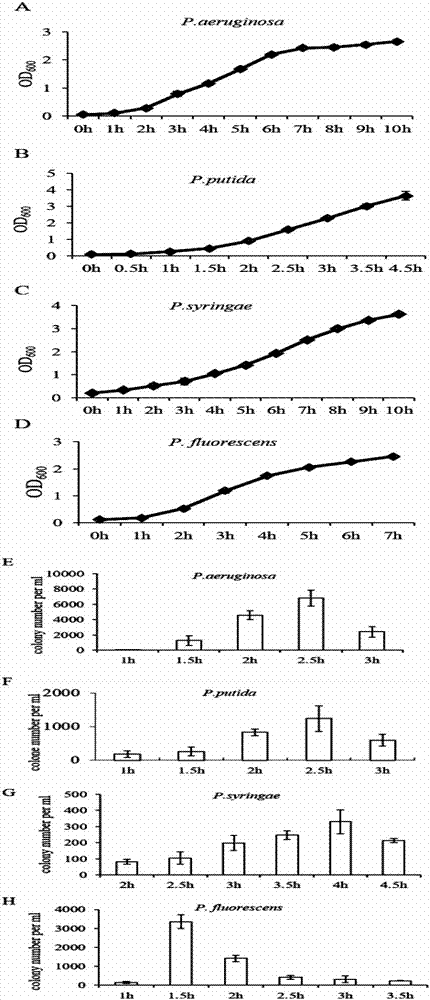
[0054] Various expression plasmids were transformed into Escherichia coli GB05 from GeneBridges in Germany, Pseudomonas aeruginosa PAO1 from DSMZ in Germany (Accession No.: DSM 1707), Pseudomonas fluorescens pf5 from American Type Culture Collection ATCC (Accession No.: BAA-477 TM ), Pseudomonas syringae DC3000 were purchased from the American Type Culture Collection ATCC (preservation number: BAA-871 TM ), Pseudomonas putida KT2440 were purchased from DSMZ, Germany (Accession No.: DSM6125). Escherichia coli competent cells were prepared according to the procedures reported in the previous literature. Pseudomonas aeruginosa, Pseudomonas fluorescens, Pseudomonas syringae, Pseudomonas putida, and strains with expression plasmids were first cultured overnight and then diluted to 1.3 ml of LB medium (Pseudomonas syringae In KB culture medium), the initial OD 600 The value of is about 0.085. Cultivate accordin...
Embodiment 1
[0055] Example 1: Construction of a series of homologous recombination expression plasmids
[0056] Using the amino acid sequence of λBeta or rac RecT as a reference, BLAST analyzes the genomes from Pseudomonas and Pseudomonas phages, looking for proteins with potential recombination functions. Two operons containing paired exonuclease-recombinase were found from Pseudomonas and Pseudomonas phage, one operon is RecTEpsy from Pseudomonas syringae, which has been successfully used in Construction of recombinant system; the second operon comes from Pseudomonas phage vB_PaeP_Tr60_Ab31, this operon encodes three proteins: Orf38 (B) contains 177 amino acids, and the sequence has 62% homology with Redβ; Orf37 (A) It contains 195 amino acids, and its sequence has 32% homology with Redα; Orf36(S) contains 148 amino acids, and its sequence has 54% homology with the single-stranded DNA binding protein SSB of Escherichia coli.
[0057] A series of recombinant functional plasmids are cons...
Embodiment 2
[0061] Example 2: Efficiency Comparison of Different Recombination Systems in Pseudomonas
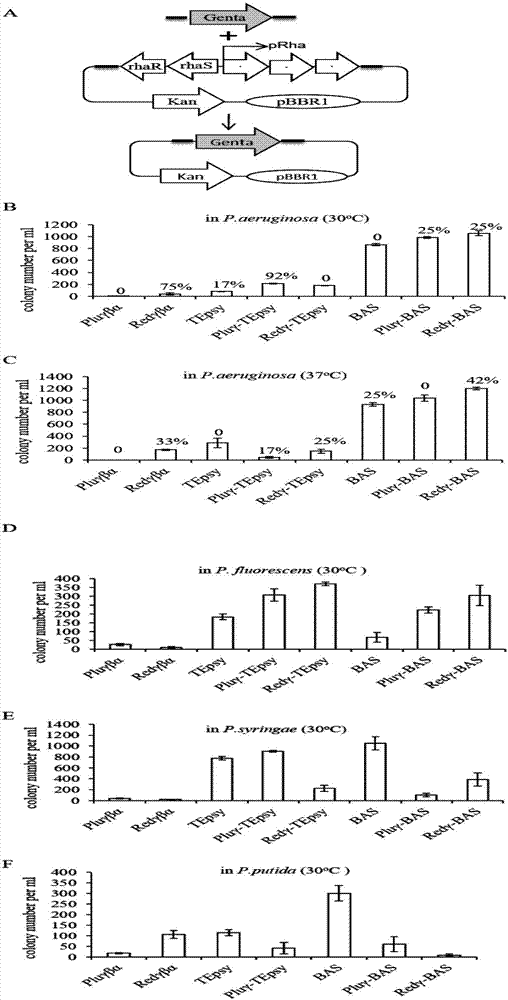
[0062] pBBR1-Rha-red_gba-Kan, pBBR1-Rha-plu_gba-kan, pBBR1-Rha-TEpsy-kan, pBBR1-Rha-pluGam-TEpsy-kan, pBBR1-Rha-redGam-TEpsy-kan, pBBR1-Rha-BAS -kan, pBBR1-Rha-pluGam-BAS-kan, pBBR1-Rha-RedGam-BAS-kan were transformed into four kinds of Pseudomonas (Pseudomonas aeruginosa, Pseudomonas syringae, Pseudomonas fluorescens) bacteria and Pseudomonas putida). figure 1 A is a schematic diagram of the comparison of recombinant system efficiencies. Pseudomonas aeruginosa grows well at both 37°C and 30°C, so a comparison of efficiencies at these two temperatures is needed. Pseudomonas aeruginosa itself has multiple drug resistances and it is easy to obtain stronger drug resistance mechanisms, so the correct rate should be calculated after comparing the recombination efficiency in Pseudomonas aeruginosa.
[0063] In Pseudomonas aeruginosa, the addition of Redγ and Pluγ proteins to the BAS recomb...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com