Method, oligonucleotide and kit for detecting high-risk HPV (human papilloma viruses)
An oligonucleotide and kit technology, applied in the field of detection of high-risk HPV, oligonucleotides and kits, can solve the problems of inability to type, increase the amount of samples and detection reagents, and increase the cost of detection, etc. The effect of ensuring the detection accuracy, reducing the background fluorescence value, and reducing the detection cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Embodiment 1: sample DNA extraction
[0047] For the sake of illustration, this embodiment takes a cervical cotton swab sample and the sample nucleic acid extraction reagent used to contain lysate, isopropanol and 1×TE buffer as examples:
[0048] (1) Take out the cervical cotton swab sample, vortex and mix well, pipette 1mL sample into a 1.5mL centrifuge tube, centrifuge at 12000rpm for 1min, carefully discard the supernatant, and keep the precipitate;
[0049] (2) Add 500 μl of lysate to the precipitate in (1), shake and mix well, and put it in a water bath or dry bath at 100°C for 10 minutes, wherein the lysate contains 0.2M NaCl, 10mM NaOH, 0.1% SDS, 0.5% NP-40 and 0.5% Tween 1×TE buffer at -20;
[0050] (3) Add 500 μl of isopropanol, vortex and mix, place at room temperature for 2 minutes, centrifuge at 12000 rpm for 5 minutes, discard the supernatant, and place at room temperature for 2 minutes;
[0051] (4) Add 50 μl of 1×TE buffer solution, fully dissolve, let...
Embodiment 2
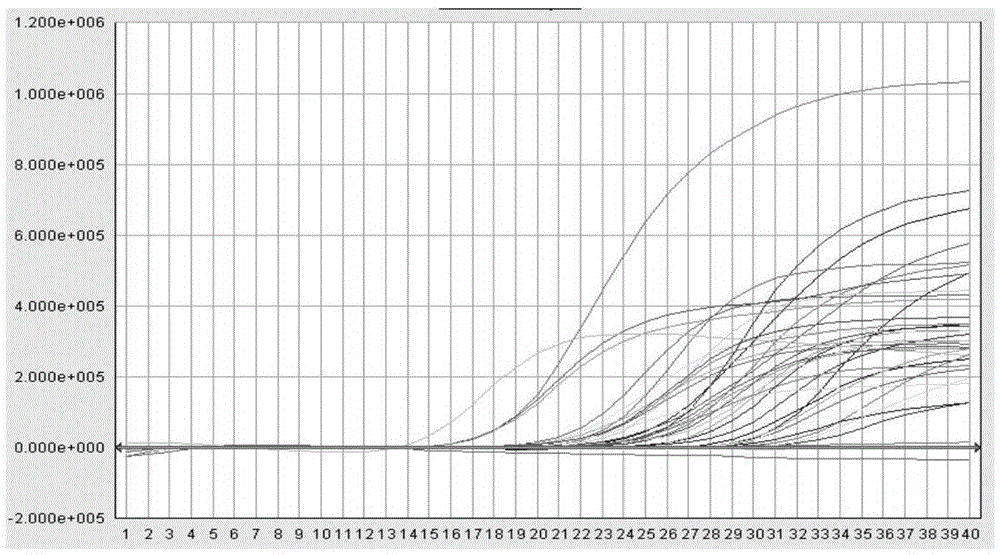
[0052] Embodiment 2: fluorescent PCR reaction steps
[0053] (1) Prepare high-risk HPV fluorescent PCR reaction solution: 4μl 10×PCRBuffer (100mM Tris-HCl, 500mM KCl); 0.08μldATP (0.4mM), 0.08μldTTP (0.4mM), 0.08μldCTP (0.4mM), 0.08μldGTP (0.4mM) ; 0.8 μl BSA (10 mg / mL); 6.4 μl MgCl 2 (25mM);0.05μlHPV16F(100μM)、0.08μlHPV16F(100μM)、0.08μlProbe1(100μM);0.05μlHPV18F(100μM)、0.08μlHPV18F(100μM)、0.08μlProbe2(100μM);0.08μlHPV31F(100μM)、0.08μlHPV35F(100μM ), 0.08 μl HPV31 / 35R (100 μM), 0.08 μl HPV33F (100 μM), 0.08 μl HPV52F (100 μM), 0.08 μl HPV58F (100 μM), 0.08 μl HPV33 / 52 / 58R (100 μM), 0.06 μl Probe3 (100 μM), 0.06 μl F55 μ M (100 μM), HPV4008 μ M); 0.06μlHPV45R(100μM)、0.08μlHPV59F(100μM)、0.06μlHPV59R(100μM)、0.06μlProbe4(100μM);0.08μlHPV39 / 68F(100μM)、0.08μlHPV39 / 68R(100μM)、0.06μlProbe5(100μM);0.08μlHPV53F(100μM )、0.08μlHPV53R(100μM)、0.08μlHPV56 / 66F(100μM)、0.08μlHPV56R(100μM)、0.08μlHPV66R(100μM)、0.06μlProbe6(100μM);0.08μlHPV26F(100μM)、0.06μlHPV51 / 82F(100μM)、0.08μlHPV26 / 51 / 82R (...
Embodiment 3
[0061] Embodiment 3: clinical sample detection
[0062] According to the method in Example 1, the DNA in 140 cases of HPV cervical swab samples was extracted, and then the DNA in the 140 cases of HPV cervical swab samples was subjected to fluorescent PCR amplification according to the method in Example 2. At the same time, for the convenience of comparison, the Roche Combas 48014 high-risk HPV (ie HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68 and 66) detection kits were used to detect the 140 The DNA in the HPV cervical swab sample was detected, and the detection results are shown in Table 4.
[0063] Table 4 Test results
[0064]
[0065]
[0066] The results in Table 4 show that among these 140 clinical samples, the present invention is consistent with the detection results of 134 clinical samples in the Roche kit, but there are also 6 clinical samples, and the present invention can detect the high-risk type The presence of HPV, while the Roche kit cannot det...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com