Cellobiose hydrolase mutant
A cellobiose and mutant technology, applied in the field of enzyme engineering, can solve the problems of high fermentation cost, large enzyme consumption, limited application of lignocellulose, etc., and achieve the effects of improving saccharification rate and strong acid resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Example 1: Screening of cellobiohydrolase mutant genes
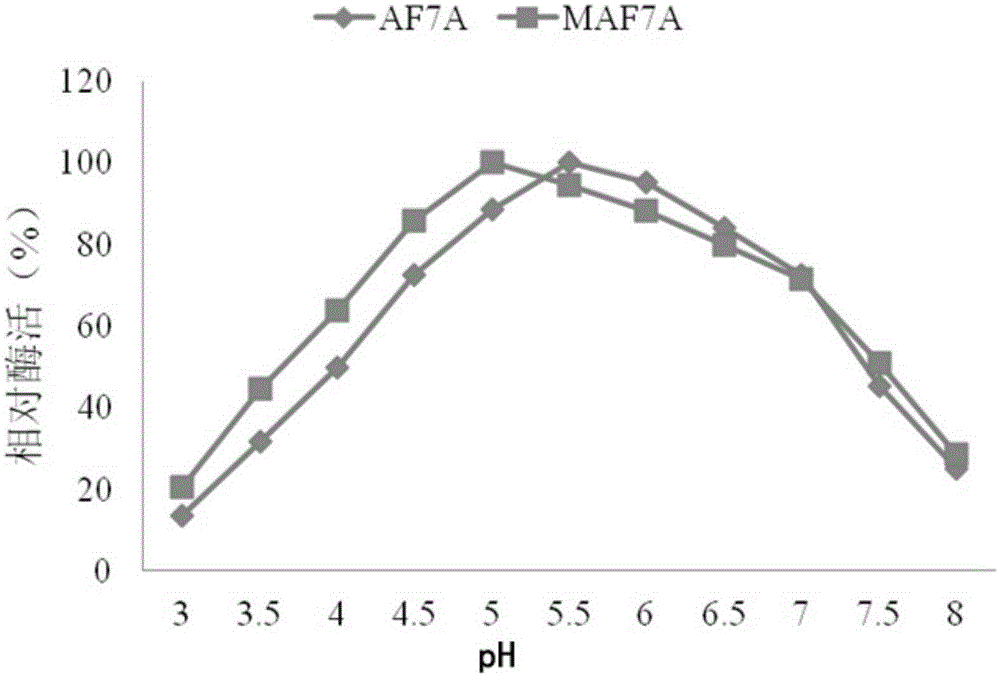
[0019] In order to improve the enzyme activity of wild-type cellobiohydrolase AF7A (the amino acid sequence is SEQIDNO: 1, the coding nucleotide sequence is SEQIDNO: 2, synthesized by Shanghai Sangon Bioengineering Co., Ltd.) under acidic conditions, through directed evolution Technology has carried out a large number of mutation screening of the enzyme, and designed PCR primers AF7A-F1 and AF7A-R1 as follows:
[0020] AF7A-F1: GGC GAATTC ATGATGCTGGCCTCCACCTTCTCC (the underline is the restriction endonuclease EcoRI recognition site);
[0021] AF7A-R1: ATAGCGGCCGCCTACAGGCACTGAGAGTAATAATC (the underline is the restriction enzyme NotI recognition site);
[0022] With the wild-type cellobiohydrolase AF7A gene SEQIDNO: 2 as template, utilize above-mentioned primer to carry out PCR amplification with GeneMorphII random mutation PCR kit (Stratagene), glue recovers PCR product, EcoRI, NotI carry out enzymatic treatment...
Embodiment 2
[0027] Example 2: Construction and verification of cellobiohydrolase mutant recombinant strains
[0028] (1) Protoplast preparation
[0029] Take the spore suspension of Trichoderma reesei (Trichodermareesei) SCHD4 strain, inoculate it on a PDA plate, and cultivate it at 30°C for 6 days; Powder, 1% glucose, 0.1% uridine) liquid medium, 30 ° C, 220rpm shaking culture for 14 ~ 16h;
[0030] Collect the mycelium by filtration with sterile gauze, and wash once with sterile water; place the mycelium in a conical flask containing 20mL of 10mg / mL lyase solution (SigmaL1412), 30°C, 90rpm for 1-2h; observe with a microscope Detection of protoplast transformation progress;
[0031] Pre-cooled 20mL1.2M sorbitol (1.2M sorbitol, 50mM Tris-Cl, 50mM CaCl 2) into the above Erlenmeyer flask, shake gently, collect the filtrate by filtering with sterile Miracloth filter cloth, centrifuge at 3000rpm at 4°C for 10min; discard the supernatant, add pre-cooled 5mL1.2M sorbitol solution to suspend ...
Embodiment 3
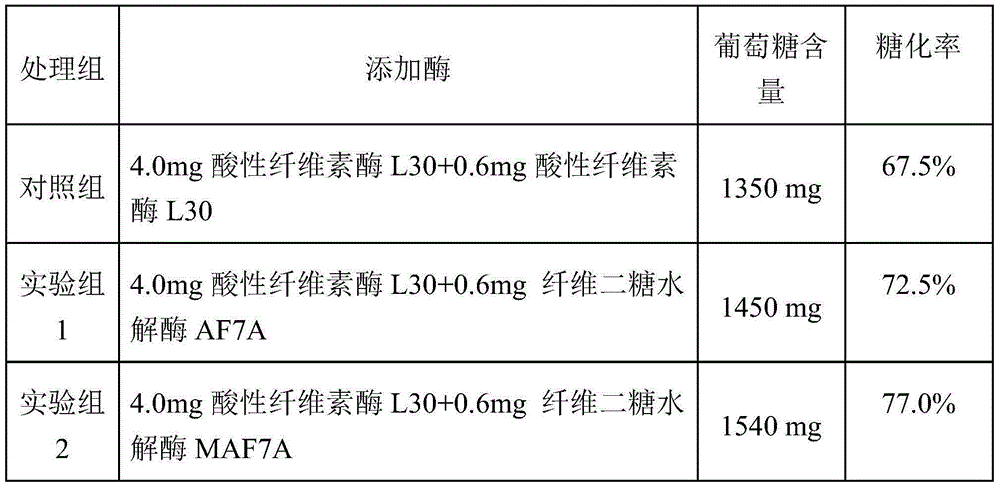
[0040] Embodiment 3 fermentation verification and enzyme activity assay
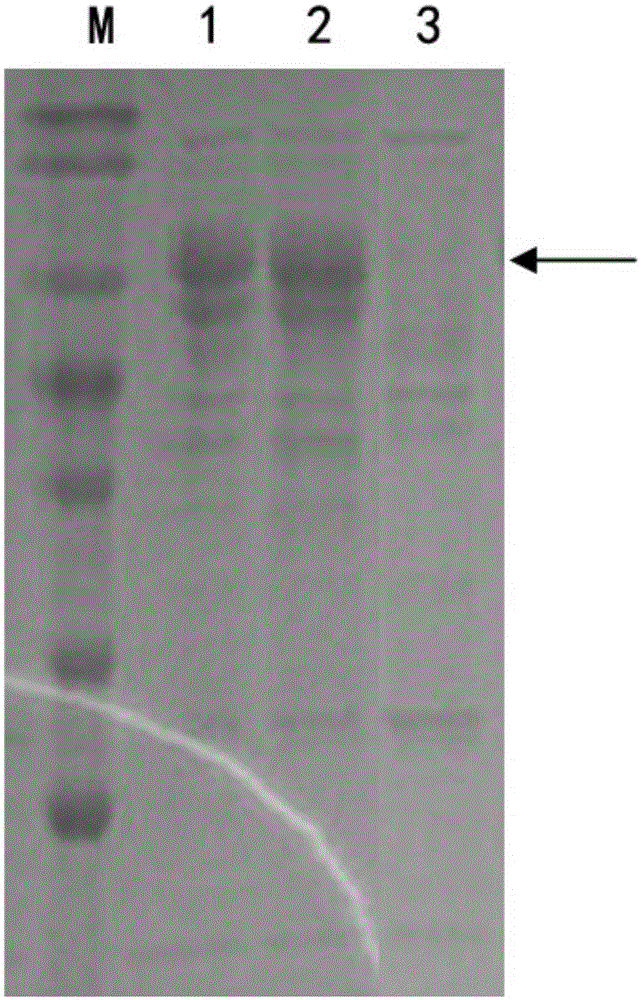
[0041] Inoculate the above-mentioned Trichoderma reesei engineering bacteria MAF7A (Trichoderma reesei MAF7A) and Trichoderma reesei engineering bacteria AF7A (Trichoderma reesei AF7A) on a PDA plate, culture at 30°C for 1 day, and after the spores are abundant, take two pieces of hyphae with a diameter of 1 cm to inoculate In 50mL fermentation medium containing (1.5% glucose, 1.7% lactose, 2.5% corn steep liquor, 0.44% (NH 4 ) 2 SO 4 , 0.09% MgSO 4 , 2% KH 2 PO 4 , 0.04% CaCl 2 , 0.018% Tween-80, 0.018% trace elements) in a 250mL Erlenmeyer flask, cultivated at 30°C for 48 hours, then cultivated at 25°C for 48 hours, and took the fermentation supernatant for SDS-PAGE electrophoresis detection and analysis. The result is as figure 1 As shown, the protein bands at 70kDa indicated by arrows in swimming lanes 1 and 2 are cellobiohydrolase AF7A and mutant MAF7A respectively, thereby illustrating that ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com