Multi-PCR detection primers for detecting four sheep pathogenic bacteria and detection method
A detection method and technology for pathogenic bacteria, applied in the field of pathogen detection, can solve the problems of low sensitivity and long detection period, and achieve the effects of high sensitivity, simple operation and good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Embodiment 1 single multiplex PCR reaction
[0063] Klebsiella pneumoniae, Proteus mirabilis, Escherichia coli and Salmonella cultured overnight were used to extract bacterial genomic DNA using a bacterial genomic DNA extraction kit (TIANGEN Company) for PCR amplification. Wherein said Klebsiella pneumoniae is selected from Klebsiella pneumoniae ATCC700603, Salmonella is selected from ATCC14028 Salmonella typhi, Escherichia coli is selected from ATCC8739 Escherichia coli, and Proteus mirabilis is selected from ATCC12453 Proteus mirabilis.
[0064] Single-plex PCR amplification reaction system (50 μL): 5.0 μL containing Mg 2+ 10×PCRbuffer[750mmol / LTris-HCl(pH8.8), 200mmol / L(NH4) 2 SO 4 , 0.1% Tween20, 25mmol / LMgCl 2 ], 5.0 μL of 2.5 mmol / L dNTP, 0.5 μL of 5 U / μL Taq polymerase, 1 μL of upstream and downstream primers (each primer concentration is 25 pmol / μL), and make up to 50 μL with water.
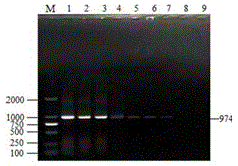
[0065] The reaction conditions were: pre-denaturation at 94°C for 7 minu...
Embodiment 2
[0068] Embodiment 2 multiplex PCR reaction
[0069] Each of the test bacteria was cultured overnight, and the bacterial genomic DNA was extracted using a bacterial genomic DNA extraction kit (TIANGEN Company) and used for PCR amplification. Among the various test bacteria adopted, wherein said Klebsiella pneumoniae is selected from Klebsiella pneumoniae ATCC700603, Salmonella is selected from ATCC14028 Salmonella typhi, Escherichia coli is selected from ATCC8739 Escherichia coli, Proteus mirabilis is selected from From ATCC 12453 Proteus mirabilis. Other bacteria E is Proteus vulgaris ATCC49132, F is Staphylococcus aureus ATCC6538, G is Listeria monocytogenes ATCC19114, and H is Pseudomonas aeruginosa ATCC9027.
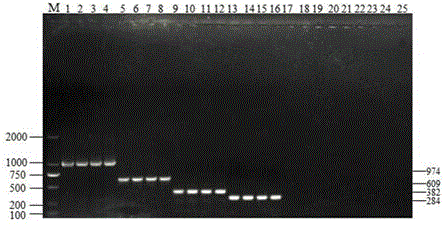
[0070] The primers were 4 pairs of specific primers designed using Klebsiella pneumoniae type III pilus structural gene, Proteus mirabilis urease synthesis positive regulator R gene, Salmonella invasive antigen conserved gene, and Escherichia coli alkaline phosphat...
Embodiment 3
[0088] Embodiment 3 clinical detection
[0089] Randomly select 20 copies of preserved sheep disease materials, carry out enrichment culture to the tissue fluid in broth culture medium respectively, through enrichment culture, the content of each pathogenic bacteria isolated is all within 10 6 cfu / mL or more. Bacterial genomic DNA was extracted, and the method in Example 2 was used for multiplex PCR amplification. At the same time, routine bacterial isolation and identification were carried out as a control.
[0090] The results of multiplex PCR amplification showed that 13 samples amplified a band of about 600bp, which showed that it contained Klebsiella pneumoniae, and 10 samples amplified a band of about 380bp, which showed that it contained Proteus mirabilis; 8 samples amplified A band of about 974bp was found, indicating that it contained Escherichia coli; 8 samples amplified a band of about 280bp, indicating that it contained Salmonella. From the analysis of the res...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com