FGFR3 single-chain antibody-protamine fusion protein and application thereof
A fusion protein and single-chain antibody technology, applied in the fields of genetic engineering and biopharmaceuticals, can solve problems such as the inability to fundamentally eradicate tumors, achieve development and in vivo safety assurance, increase protein expression, and facilitate separation and purification.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] PCR synthesis of the R3‐P fusion gene.
[0045] According to the known FGFR3 monoclonal antibody sequence, we conducted bioinformatics analysis to find out its possible heavy chain and light chain sequences, and then compared and analyzed multiple reported FGFR3 ScFv sequences, according to the high consensus rate (>50%) codons, point mutations were carried out, and Cys (Gly was mutated to Cys) was introduced into the non-critical regions of the heavy chain and light chain at the same time, so as to facilitate the formation of disulfide bonds in the chain without affecting its binding function. At the same time to enhance the stability of the single chain antibody. The assembly sequence of each part is as follows figure 1 shown.
[0046] The nucleotide sequence and coding sequence of the entrusted company to synthesize the R3-fusion gene are shown in Table Seq ID No.2 and Seq ID No.1, respectively.
Embodiment 2
[0048] Construction of pET‐20b‐R3‐P expression vector.
[0049] In order to subclone the R3P gene into pET-20b, the ScFv-protamine (R3-P) sequence was synthesized by PCR using P1 and P2 as primers (see Table 1 for primer sequences).
[0050] PCR reaction system:
[0051]
[0052]
[0053] Reaction program: pre-denaturation at 98°C for 5min; denaturation at 98°C for 10min; annealing at 55°C for 15s; extension at 72°C for 1min, 30 cycles, and finally extension at 72°C for 5min. After no non-specific bands were detected by 1% agarose gel electrophoresis, the product DNA fragments were purified and recovered with a DNA gel recovery kit. The result is as figure 2 shown. Then the fragment was double-enzyme-digested with Nde Ⅰ and Xho Ⅰ, and carried out an enzyme ligation reaction with the pET-20b plasmid that was also double-digested with the two enzymes to obtain the pET-20b-R3-P plasmid.
[0054] Table 1 Primer Sequence
[0055] Primer Sequence (3'→5') ...
Embodiment 3
[0057] Obtaining of R3‐P fusion protein.
[0058] 1. Induced expression
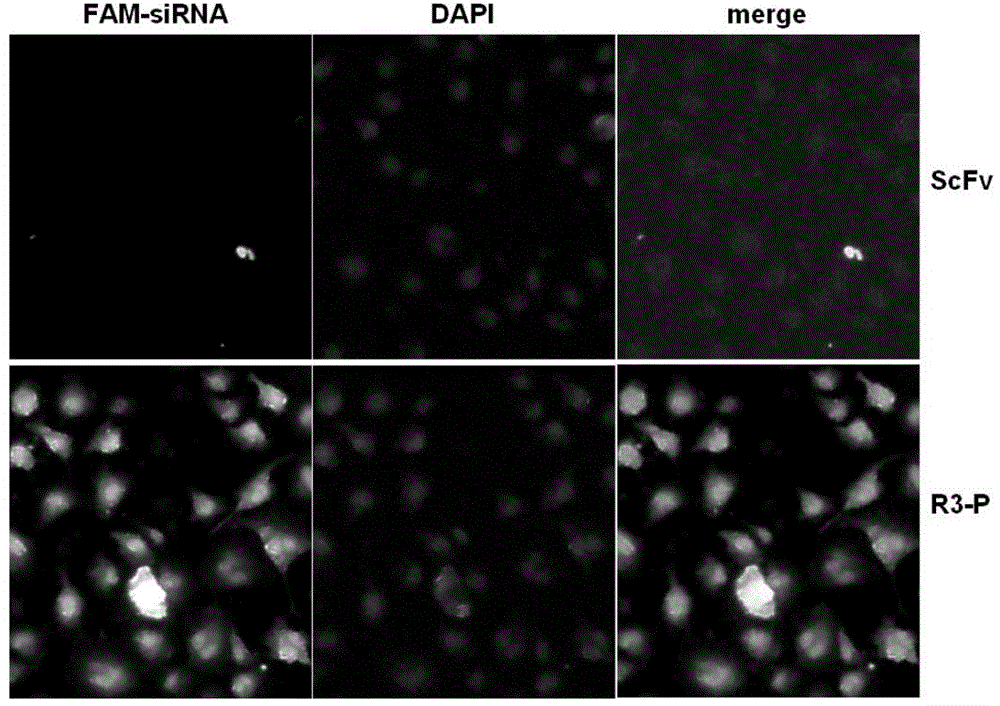
[0059] The correctly sequenced pET‐20b‐R3‐P plasmid was transformed into Escherichia coli BL21 (DE3) competent cells, and positive transformants were screened on ampicillin resistance plates. A single colony of the screened positive recombinant bacteria BL21(DE3)pET‐20b‐R3‐P was inoculated into LB medium and cultured with shaking at 37°C for 16 hours. Then it was transferred to fresh LB medium at a ratio of 5%, and the expression conditions such as temperature, IPTG concentration and induction time were optimized respectively. The results showed that when the OD value was 0.6, induced by 1mM IPTG at 37°C for 3h, ultrasonic (200w, ultrasonic 5s, interval 5s, cycle 180 times) destroyed the bacteria, and the target protein expressed in the form of inclusion bodies could account for the total bacterial cells. More than 85% of the target protein, the results are as follows image 3 shown.
[0060] 2. Prot...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com