A set of lamp detection primers and kits for microsporidia in silkworm eggs
A technology of silkworm egg microspores and detection kits, applied in the biological field, can solve the problems of silkworm egg DNA detection interference, complicated separation and collection of microsporidia, and many operating steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0091] Example 1 Primer Design
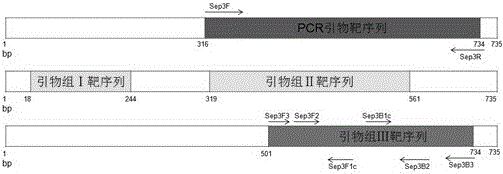
[0092] 1. According to the sequence of the No. silkworm septin3 gene (Accession number: KJ451482.1) verified by the laboratory through the transcriptome sequencing method and the clone sequencing method, the homology analysis was carried out by BLAST software, and no similar sequence was found. The invention designed a set of PCR primers with this gene as the target gene: primer Sep3F / Sep3R.
[0093] The sequence of primer Sep3F (shown in SEQ ID NO:5) is:
[0094] 5' TTCGGAGTAATAGCCAGTG 3'
[0095] The sequence of primer Sep3R (shown in SEQ ID NO:6) is:
[0096] 5'-AATTTGTTACAGAAGGACTCC-3'
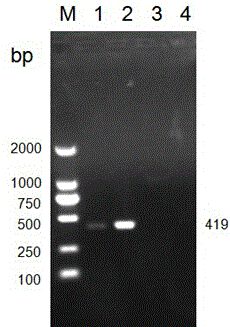
[0097] 2. Use the above-mentioned PCR primers to detect healthy silkworm eggs and silkworm eggs infected with N. figure 1 shown).
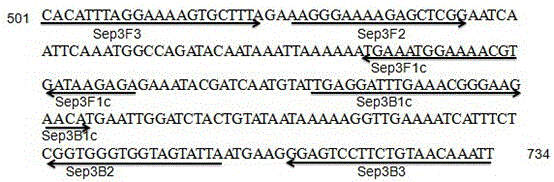
[0098] Therefore, based on the position of the amplified fragment of the PCR primer, using the septin3 gene as the target gene designed by the LAMP primer, use the online software Primer ExplorerV4 (http: / / primerexplorer.j...
Embodiment 2
[0118] Embodiment 2 kit preparation
[0119] 1. For the preparation of DNA template of No. spp. silkworm, QIAGEN Company's plant mini-extraction kit was used, and the method was carried out according to the instructions. Specifically include the following steps:
[0120] S1. Take 200 μL microsporidia sample, centrifuge at 12,000 rpm for 5 minutes, discard the supernatant; then add 50-100 μL ddH 2 O resuspension;
[0121] S2. Draw the resuspended spore liquid into the pre-cooled mortar, and grind it fully with liquid nitrogen (more than 3 times);
[0122] S3. Put the ground spores into a 1.5 mL centrifuge tube, add 400 μL lysis buffer AP1 and 4 μL RNase A, and vortex to mix (400 μL lysis buffer AP1 and 4 μL RNase A should not be mixed before use);
[0123] S4. Incubate the mixed solution at 65°C for 10 min (invert the test tube 2 to 3 times during the period);
[0124] S5. Add 130 μL buffer P3, mix and ice-bath for 5 minutes; then centrifuge at 14,000 rpm for 5 minutes;
...
Embodiment 3
[0142] Example 3 Kit detection method
[0143] 1. The LAMP reaction conditions of the kit are: constant temperature reaction at 63°C for 30-60 minutes; then inactivation at 95°C for 2 minutes.
[0144] 2. When staining or agarose gel electrophoresis is used to determine the test result, the reaction system of the kit is:
[0145] 2×Reaction Buffer 12.5μL
[0146] Primers Sep3F3 / Sep3B3 and Sep3FIP / Sep3BIP 1 μL
[0147] Bst DNA polymerase 8U / μL 1μL
[0148] Template DNA 2 μL
[0149] wxya 2 O 8.5 μL;
[0150] When the detection result is judged by the real-time fluorescence method, the reaction system is:
[0151] 2×Reaction Buffer 12.5μL
[0152] Primers Sep3F3 / Sep3B3 and Sep3FIP / Sep3BIP 1 μL
[0153] Bst DNA polymerase 8U / μL 1μL
[0154] Fluorescent indicator 0.5μL
[0155] Template DNA 2 μL
[0156] wxya 2 O 8 μL;
[0157] Wherein, the concentration of the primer Sep3F3 / Sep3B3 is 5 pmol / μL, and the concentration of the primer Sep3FIP / Sep3BIP is 20˜40 pmol / μL.
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com