A method for detecting locust microsporidiosis by pcr
A technology for locust microsporidia and locusts, applied in the field of PCR detection of locust microsporidiosis, can solve the problems of no rapid detection method of molecular biology, observation of morphological differences, time-consuming and labor-intensive sensitivity, etc., to reduce detection costs and workload, improved detection accuracy, and reduced sample volume
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] The extraction of embodiment 1 locust microsporidia sample DNA
[0028] Get locust microsporidia, adopt CTAB method to extract DNA, specifically:
[0029] Take 1ml of Microsporidia locust and centrifuge to remove the supernatant. Grind the sample with electric tissue grinding rod for 20-30s; add 0.5ml CTAB buffer solution to each 1.5ml centrifuge tube, mix well, add 20μl of 20mg / ml proteinase K to each tube, and shake on the oscillator for 30s; 56℃ water bath for 6h; add an equal volume of phenol / Chloroform / isoamyl alcohol (volume ratio 25:24:1) for extraction once; 12000rpm, 10min; take the supernatant, add an equal volume of chloroform for extraction once; 12000rpm, 10min; take the supernatant, add an equal volume of isopropanol, Precipitate on ice for 20 minutes; 12000 rpm, 10 minutes, discard the supernatant; wash once with 500 μl of 75% ethanol, centrifuge slightly, discard the supernatant; wash once with 500 μl absolute ethanol, centrifuge slightly, discard the ...
Embodiment 2
[0030] The design of embodiment 2 primers
[0031] According to the known sequence (gene sequence number: GQ397125), design primers:
[0032] Nlos-F5: 5'-GCTCCAAATACAGCGTAAATGC-3' (SEQ ID NO: 1);
[0033] Nlos-R5: 5'-GAGATTCGCATTCGCCACAGC-3' (SEQ ID NO: 2).
Embodiment 3
[0034] The PCR amplification procedure of embodiment 3 locust microsporidia
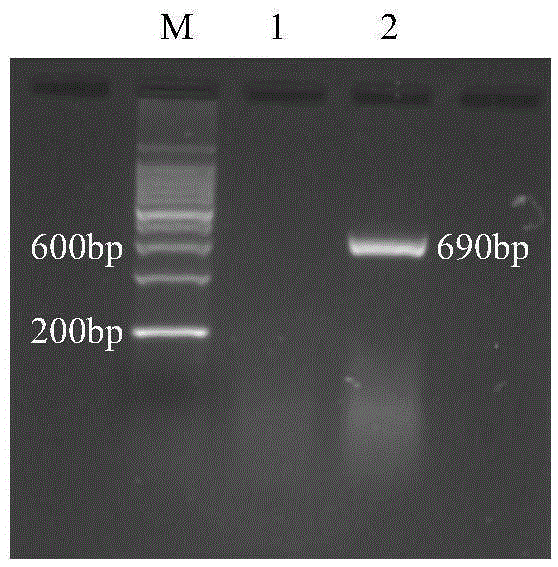
[0035] The DNA of the tested sample was used as a template, and Nlos-F5 and Nlos-R5 were used as forward and reverse primers to perform fragment amplification. The PCR reaction system is: 2×TaqMasterMix (Tiangen Biochemical Technology Co., Ltd.), 12.5 μl; 10 μM primer Nlos-F5, 1 μl; 10 μM primer Nlos-R5, 1 μl; DNA template, 1 μl; ddH 2 O, 9.5 μl; the total reaction system is 25 μl. The reaction conditions were: 95°C, 5min, 1 cycle; 94°C, 30s, 66°C, 30s, 72°C, 30s, 35 cycles; finally 72°C, 10min, 1 cycle. Take 10 μl of PCR product, electrophoresis in 1% agarose gel [containing nucleic acid dye (10000 × GeneGreen)], voltage 100V, 40min, observe the PCR diagnosis result under strong ultraviolet light (results see figure 1 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com