Method of shearing fusion protein by escherichia coli intracellular protease
A fusion protein and Escherichia coli technology, applied in the field of genetic engineering, can solve the problems of protein inactivity, expensive protease, and large economic burden
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
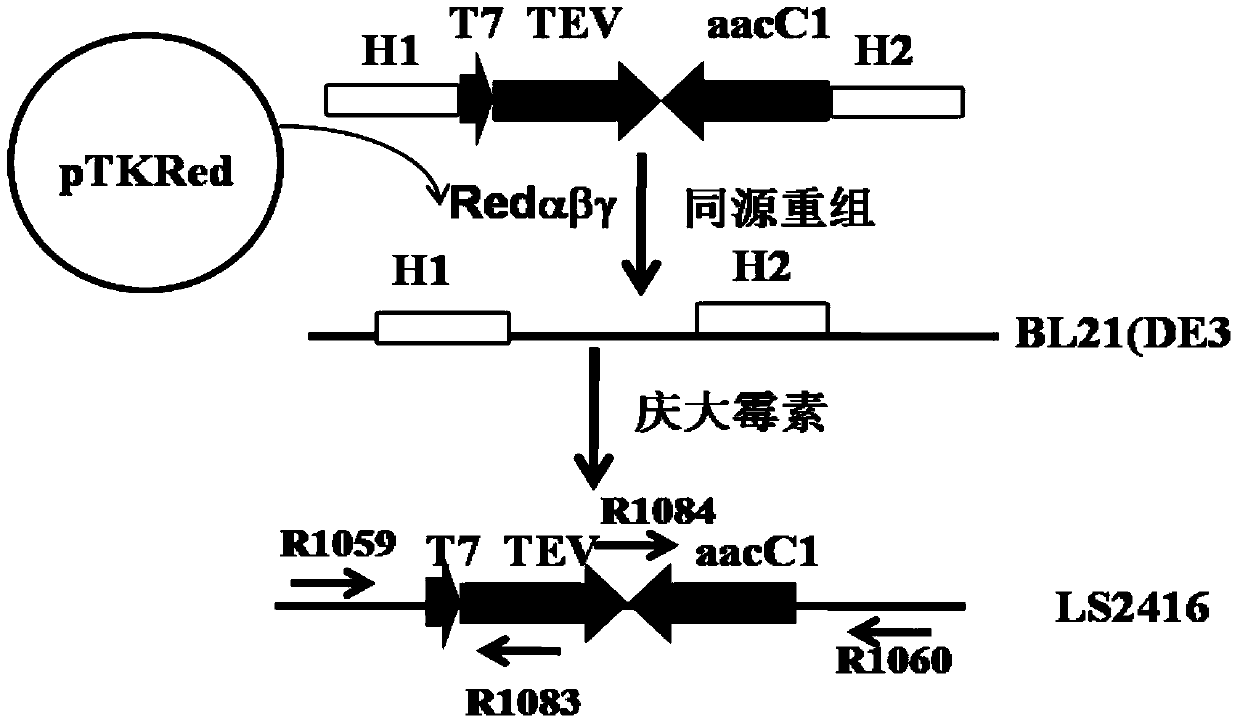
[0054] Example 1. Construction of intracellular shear strain with TEV protease gene integrated into BL21(DE3) chromosome
[0055] A vector containing a resistance gene and a homologous fragment for homologous recombination is first constructed. Design primer R1077: 5'- GTTAACGAGCTC GAATTGGCCGCGGCGTTG-3', (SEQ ID NO. 1), the restriction sites HpaI and SacI are underlined; R1078: 5'- GAGCTC GAATTGACATAAGCCTG-3', (SEQ ID NO. 2), SacI is underlined. Using pBAD322 as a template, a 0.8kb gentamycin gene fragment was obtained by PCR amplification. The 0.8kb blunt end was cloned into the SnaBI site of pLS2429 to obtain the recombinant clone pLS2459. After pLS2459 was digested with KpnI-SaII, the 1.8kb fragment was cloned into the KpnI-SaII site of pR6KMCS to obtain recombinant clone pLS2460. pLS2460 uses the R6K replicon, so the host strain is BW25141. BL21(DE3) does not contain the Pir gene required for R6K replication, so the circular vector remains without background interfe...
Embodiment 2
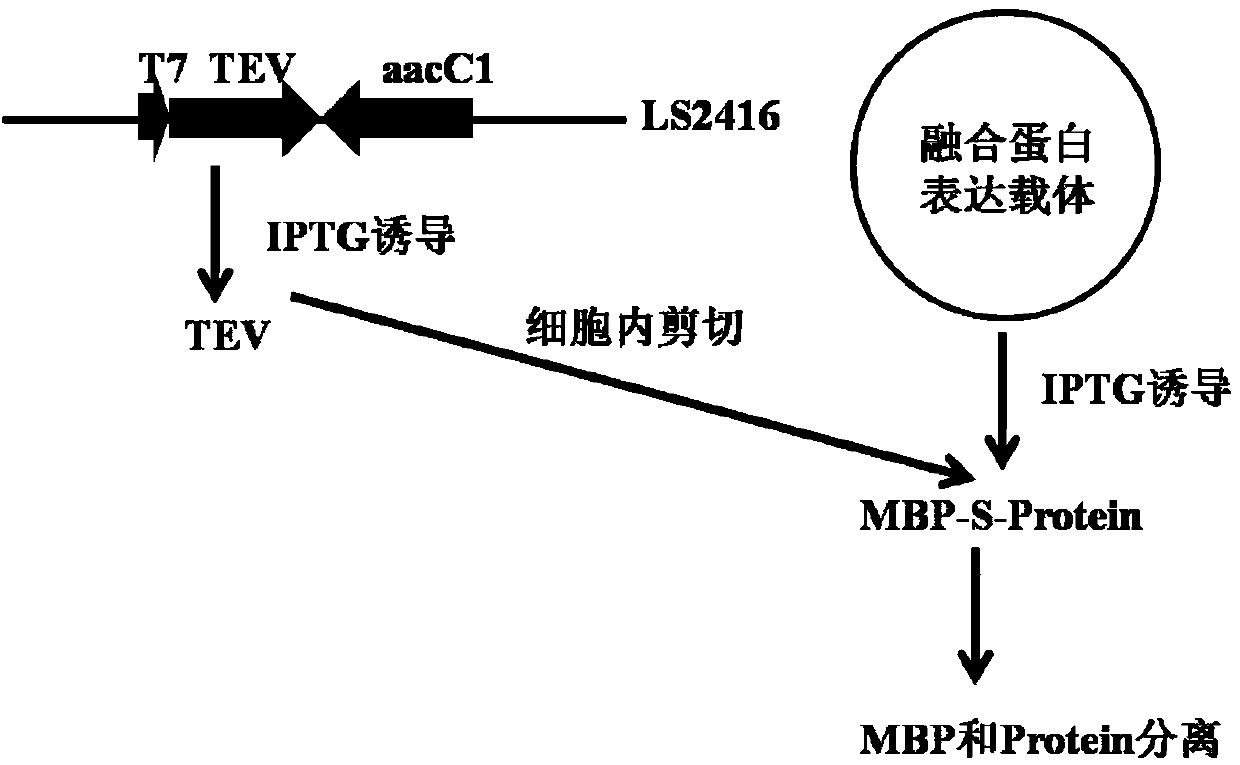
[0061] Example 2. Construction of expression vectors
[0062] pLS912 is a fusion expression vector containing two fusion tags of His and MBP and a 1.5kb stuffer fragment, described in Reference 7. To remove the His tag fused to MBP, design primer P1901: 5'-GGG GAATTCATATG GAAGAAGGTAAACTGGTAATCTG-3', (SEQ ID NO. 11), the introduced EcoRI and NdeI restriction sites are underlined; P1902: 5'-GGG GGATCC AGC AGA TCT TTGTTATAAATC-3', (SEQ ID NO. 12), the introduced EcoRI site and the BglII restriction site contained in the amplified sequence are underlined. Using pLS912 as a template, a 0.4kb fragment was obtained by PCR amplification. After being digested with NdeI-BglII, it was ligated with a vector of 7.5kb pLS912 digested with NdeI-BglII, and the recombinant clone pLS1902 was obtained by screening. GAGAATCTTTATTTTCAGGGC is contained between the MBP fusion tag and the stuffing fragment, and its amino acid sequence is ENLYFQ↓G, and ↓ indicates the TEV restriction site. ...
Embodiment 3
[0064] Reality Example 3. Intracellular cleavage of MBP-GFP protein
[0065] Design primer RG3: 5'-GAA GGATCC AGCAAGGGCGAGGAGCTGTTC-3', (SEQ ID NO. 13), RG4: 5'-GAA CTCGAG CTTGTACAGCTCGTCCATGCC-3', (SEQ ID NO. 14), the introduced BamHI and XhoI restriction sites are underlined. Using plasmid pJOE4905.1 as a template, PCR amplification of 0.7kb GFP gene was obtained. After digestion with BamHI-XhoI, it was cloned into the BamHI-XhoI digestion site of pKS, and the digestion and sequencing were correct to obtain pLS2436. pLS2436 was digested with BamHI-XhoI, and a 0.7kb GFP gene fragment was recovered and cloned into pLS1902, which was digested with BamHI-XhoI to a 6.4kb vector to obtain the MBP-GFP fusion expression vector pLS2437. The 3' end of GFP is fused to the Histag on the vector, followed by a terminator to terminate expression. pLS2437 was transformed into BL21(DE3), and the expression was induced by 1 mM IPTG at 30°C, and the 69.1KD expression protein was obtaine...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com