ShRNA (short hairpin ribonucleic acid) suppressing IRS1 (insulin receptor substrate 1) gene expression and application
A gene expression and gene technology, applied in the field of genetic engineering, can solve the problems of short lifespan of mice, inability to be used as a long-term disease research target, and inability of mice to simulate human diseases well.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
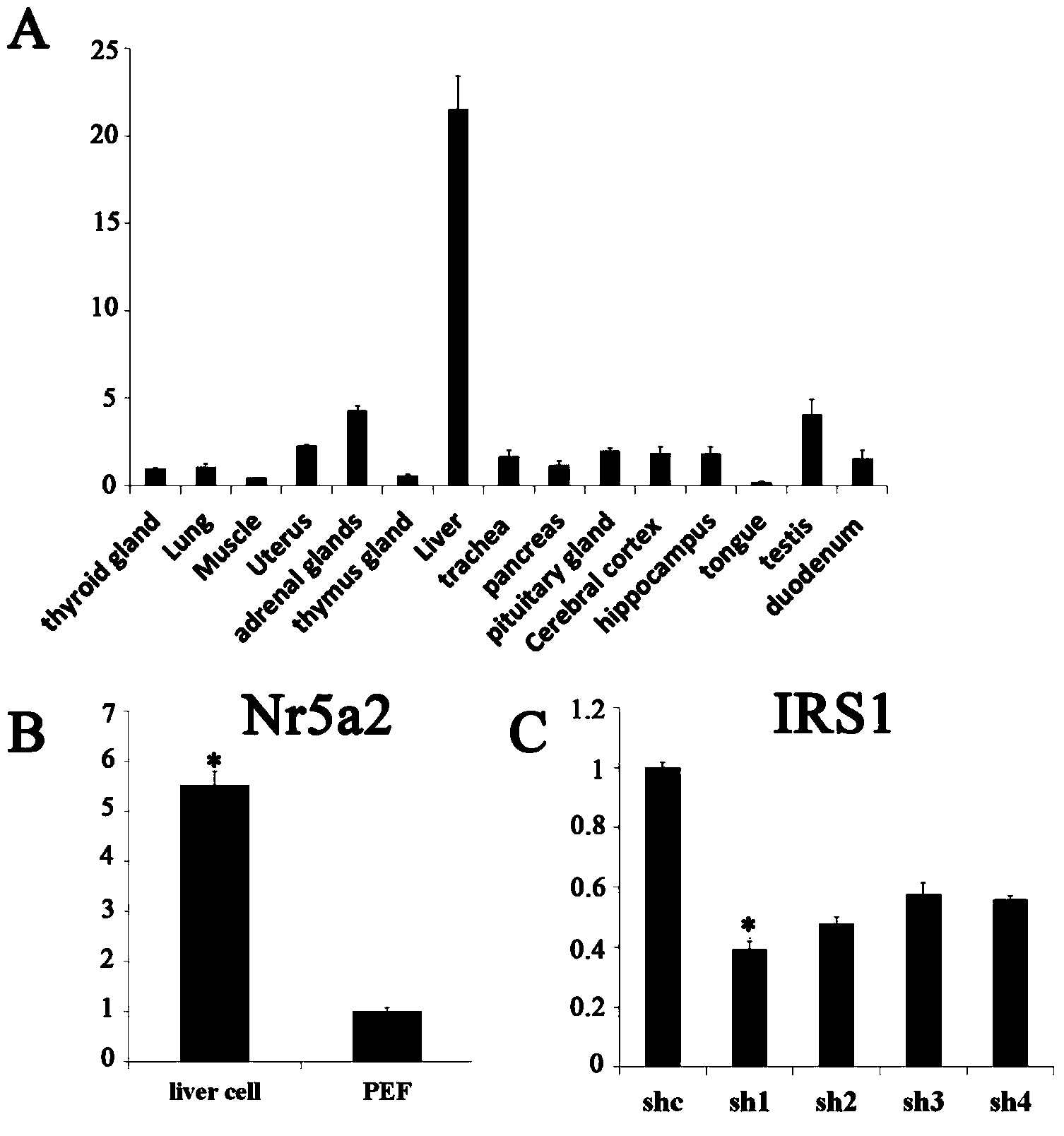
[0040] Cloning of porcine IRS gene and its expression in various tissues
[0041] (1) Cloning of porcine IRS1 gene
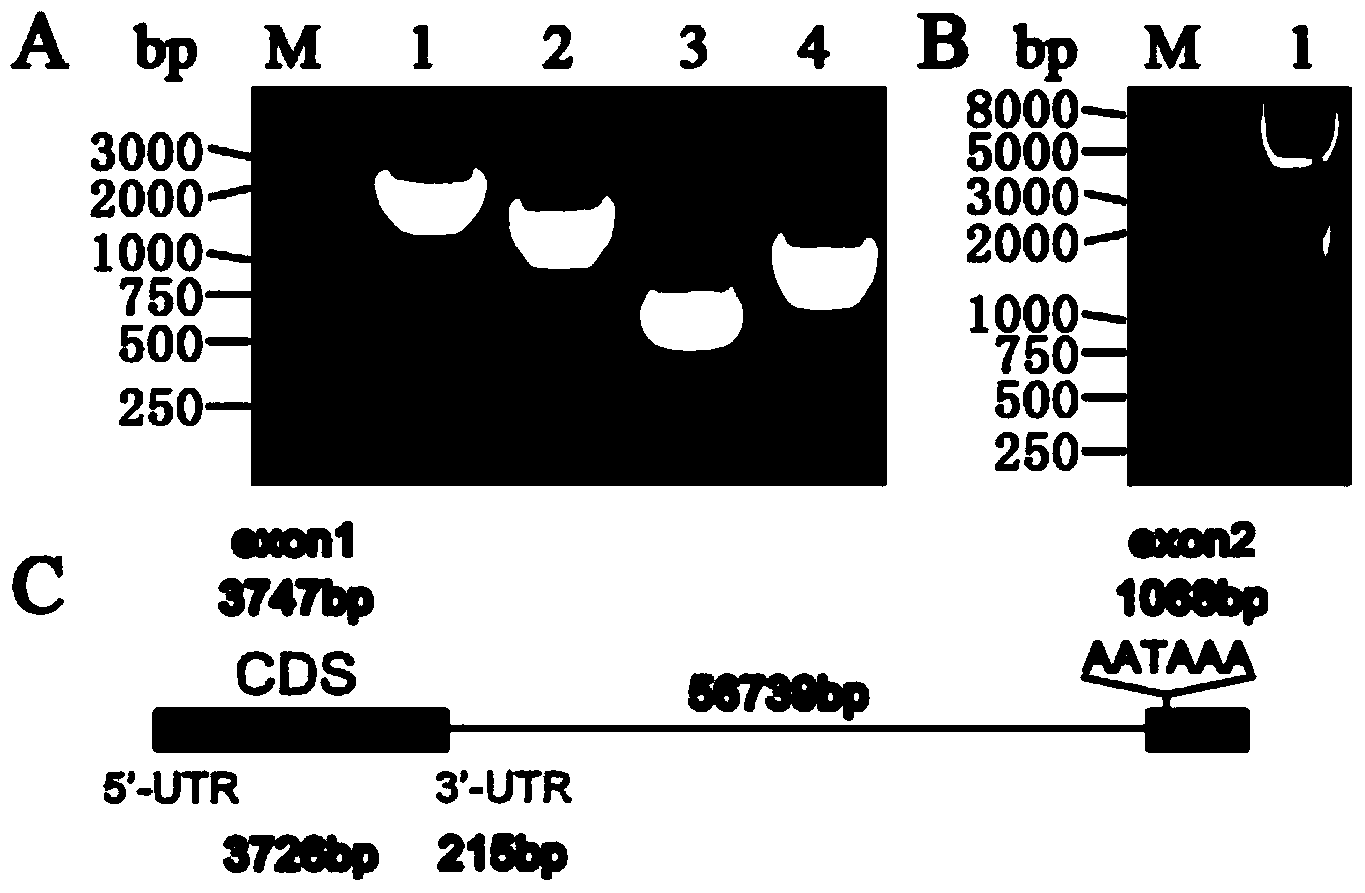
[0042] According to the known porcine IRS1 gene sequence, 4 pairs of primers (Table 1) were designed, and the full-length porcine IRS1 gene was cloned by overlapping PCR method. Among them, the 1st, 2nd, and 4th pair of primers are exon regions, and do not span introns, and PCR amplification is performed directly using the genome of Bama pigs as a template. Liver cDNA was used as a template for PCR amplification, and the total system was 50ul. The PCR reaction system includes: LA Taq 1ul, 10×LA Taq Buffer 5ul, dNTPs 4ul, template 1ul, Forward Primer 2ul, Reverse Primer 2ul, dH2O 35ul. The PCR reaction parameters were: pre-denaturation at 94°C for 5min; denaturation at 94°C for 30s, annealing at 60°C for 30s, extension at 72°C for 2min, 35 cycles; final extension at 72°C for 10min. The results of PCR products were observed in 1% agarose gel electrophoresis, an...
Embodiment 2
[0052] Construction of p-Genesil-shIRS1 / shIRS2 interference vector
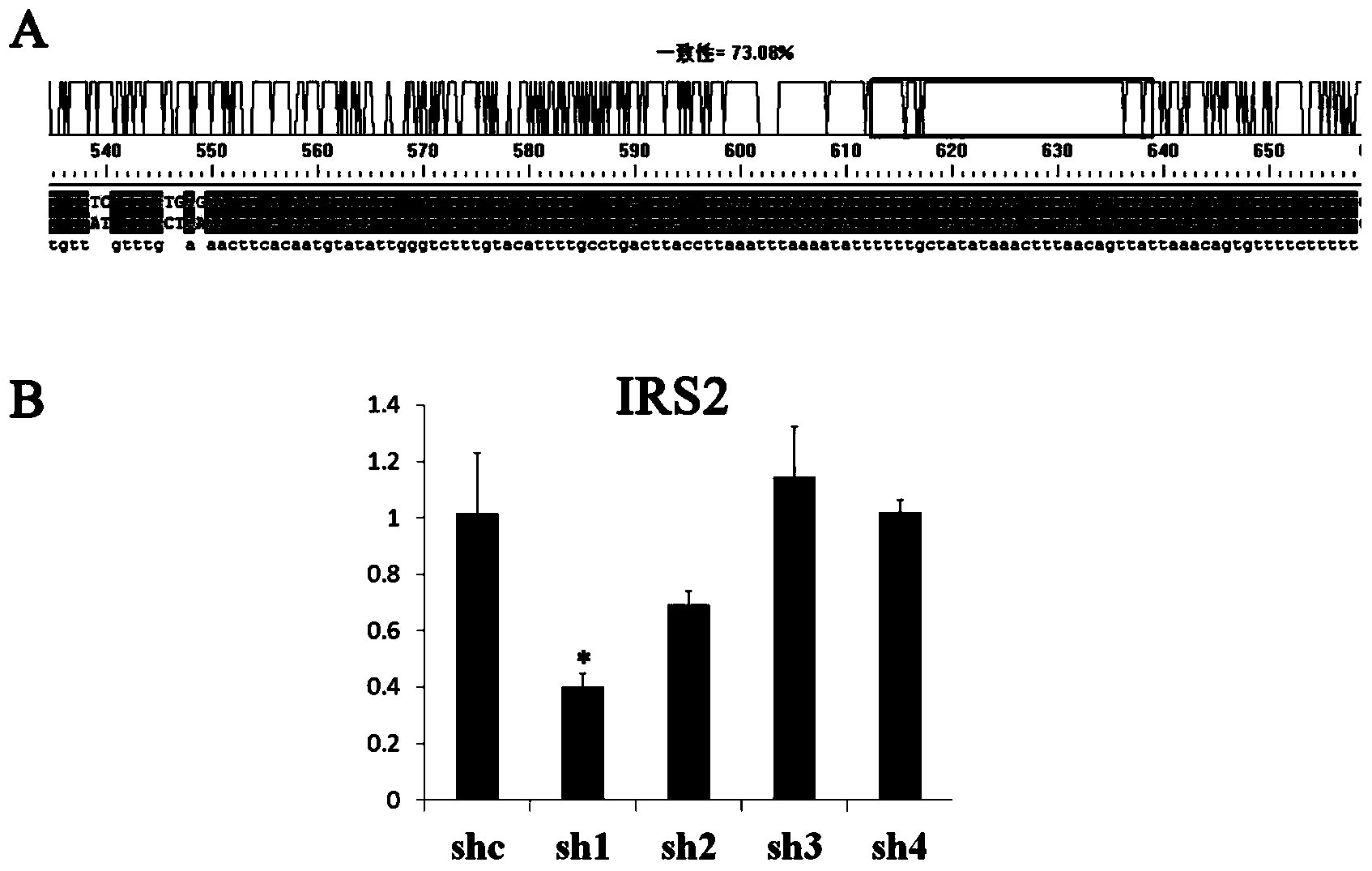
[0053] According to the porcine IRS1 and IRS2 sequences obtained by cloning, four pairs of interference fragments were designed through the Ambion website (Table 2), and single-stranded shRNA interference fragments with BamH I and Hind III restriction sites were synthesized, and after renaturation into double strands, ligation to the p-Genesil 1.0 vector linearized with BamHI and HindIII. After the sequencing was correct, the successfully constructed vectors were p-Genesil-shIRS1-1-4 and p-Genesil-shIRS2-1-4.
[0054] shIRS1-1 and shIRS2-1 are effective interference fragments screened out respectively, re-synthesized single-stranded shIRS2 interference fragments with Nhe I and Xho I restriction sites, annealed into double strands, and connected to the effective screening fragments Nhe I and Xho I linearized p-Genesil-shIRS1 vector. Afterwards, the hU6 promoter was amplified by PCR, with SnaB Ⅰ and Nhe Ⅰ res...
Embodiment 3
[0056] Cultivation of Porcine Liver Primary Cells and Detection of Interfering Carrier Effect
[0057] (1) Culture of primary porcine liver cells
[0058] Take 1d Bama pig liver tissue about 0.5cm 3Place it in physiological saline containing double antibodies, wash the tissue, peel off the mesangium on the surface of the liver, remove blood and other sundries, and wash until the liver tissue is bright red, and the lotion has no obvious sundries. After transferring the tissue to a Petri dish, cut it up with surgical scissors. Add 1ml of collagenase and place in a 37°C incubator for digestion for 5min. Add culture medium to terminate. The tissue was blown away, and the mixture was added to a centrifuge tube for centrifugation at 1200 rpm for 3 min. After centrifugation, discard the supernatant and add 3ml culture medium to resuspend. Inoculate into three large dishes and transfer to a 37°C incubator for cultivation. Observe that the cells have tissue pieces attached to the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com